Figure 5.
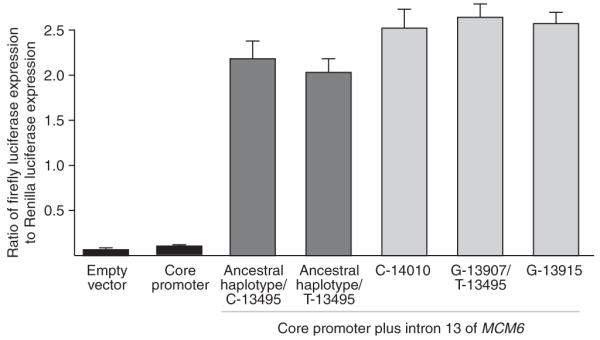
Dual-luciferase reporter assay of LCT promoter and MCM6 introns. As a control, cells were transfected with the promoterless pGL3-basic vector (‘empty vector’). Basal levels of expression were assessed using a pGL3-basic vector with 3 kb of the 5′ flanking region of LCT (‘core promoter’). Five different haplotypes of the MCM6 intron 13 were inserted upstream of the core promoter that differed at the following sites: (i) a haplotype that is ancestral for the three lactase persistence–associated SNPs, with a C at position -13495; (ii) a haplotype that is ancestral for the three lactase persistence–associated SNPs, with a T at position -13495; (iii) a haplotype that differs from (i) only at C -14010; (iv) a haplotype that differs from (i) at G-13907 and T-13495 and from (ii) only at G-13907; and (v) a haplotype that differs from (i) only at G-13915. Expression levels are reported as the ratio of firefly to Renilla luciferase; error bars represent a 95% c.i. The differences between the core promoter alone and all five MCM6 intronic constructs, as well as between the three derived versus two ancestral haplotypes, were significant (P < 0.0008, paired t tests). There was no significant difference in expression between the empty vector and the core promoter, between the two ancestral haplotypes (with and without the T-13495 allele) or between the three derived haplotypes. The construct with ancestral lactase persistence–associated alleles that differed at T-13495 served as an internal control for the expression differences for the G-13907 and T-13495 alleles, indicating that only the G-13907 allele results in increased gene expression.
