Abstract
The tumor suppressor activity of p53 is regulated by interactions with the ubiquitin ligase HDM2 and the general transcriptional coactivators CBP and p300. Using NMR spectroscopy and isothermal titration calorimetry, we have dissected the binding interactions between the N-terminal transactivation domain (TAD) of p53, the TAZ1, TAZ2, KIX, and nuclear receptor coactivator binding domains of CBP, and the p53-binding domain of HDM2. The p53 TAD contains amphipathic binding motifs within the AD1 and AD2 regions that mediate interactions with CBP and HDM2. Binding of the p53 TAD to CBP domains is dominated by interactions with AD2, although the affinity is enhanced by additional interactions with AD1. In contrast, binding of p53 TAD to HDM2 is mediated primarily by AD1. The p53 TAD can bind simultaneously to HDM2 (through AD1) and to any one of the CBP domains (through AD2) to form a ternary complex. Phosphorylation of p53 at T18 impairs binding to HDM2 and enhances affinity for the CBP KIX domain. Multisite phosphorylation of the p53 TAD at S15, T18, and S20 leads to increased affinity for the TAZ1 and KIX domains of CBP. These observations suggest a mechanism whereby HDM2 and CBP/p300 function synergistically to regulate the p53 response. In unstressed cells, CBP/p300, HDM2 and p53 form a ternary complex that promotes polyubiquitination and degradation of p53. After cellular stress and DNA damage, p53 becomes phosphorylated at T18 and other residues in the AD1 region, releases HDM2 and binds preferentially to CBP/p300, leading to stabilization and activation of p53.
Keywords: p53 transactivation domain, phosphorylation, protein–protein interaction, transcriptional coactivator, tumor suppressor
The p53 tumor suppressor is activated as a transcriptional regulator in response to DNA damage, leading to the arrest of cell growth and apoptosis. p53 is a modular protein that binds DNA as a tetramer; each subunit contains an N-terminal transactivation domain (TAD), proline-rich domain, core DNA binding domain, tetramerization domain, and C-terminal regulatory domain. In the absence of cellular stress, p53 binds target promoters in an inactive latent state and recruits HDM2 (the human homolog of mouse double minute 2, MDM2) to chromatin (1, 2). HDM2 functions as a ubiquitin E3 ligase that maintains p53 at low levels by continuous proteasomal degradation (3). DNA damage initiates a cascade of phosphorylation and acetylation events at multiple sites on p53 (Fig. 1A), resulting in stabilization and enhancement of p53 transcriptional activity (4–7). In particular, phosphorylation at threonine-18 (T18) helps stabilize p53 by inhibiting binding to HDM2 (8, 9), whereas phosphorylation of serines 15 and 20 (S15, S20) enhances recruitment of the general transcriptional coactivators and acetylases, CREB binding protein (CBP) and p300 (10–12). S15 must be phosphorylated before phosphorylation can occur at T18 and S20 (13).
Fig. 1.
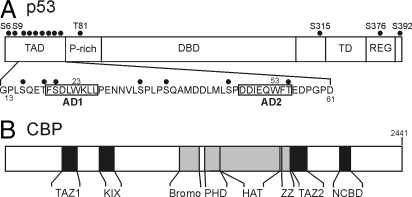
Domain organization of p53 and CBP/p300. (A) Domains of p53. TAD (N-terminal transactivation domain), P-rich (proline-rich), DBD (DNA-binding domain), TD (tetramerization domain), and REG (C-terminal regulatory domain). The location of the AD1 and AD2 motifs is indicated on a partial amino acid sequence; known sites of phosphorylation are indicated by dots. (B) Domains of CBP/p300. Domains that interact with the p53 transactivation domain, TAZ1 (residues 340–439), KIX (586–672), TAZ2 (1764–1855), NCBD (2059–2117) are shown in black.
CBP and p300 play a central role in regulation of p53 stability and the response to genotoxic stress (14–16). In unstressed cells, p53 and HDM2 form a ternary complex with the N-terminal region of CBP/p300, which promotes polyubiquitination and degradation of p53 (17, 18). After activation of p53 in response to DNA damage, HDM2 is released and p53 is stabilized and binds more tightly to CBP/p300. Direct interactions between CBP/p300 and the p53 TAD are essential for activation of transcription from p53-responsive genes. The p53 TAD (residues 1–63) contains 2 subdomains, AD1 (residues 1–40) and AD2 (residues 43–63) (19–21) and is intrinsically disordered (22, 23). Amphipathic motifs within both AD1 and AD2 form stable helical structure upon binding to target proteins (24–26).
CBP and p300 are modular proteins containing domains (Fig. 1B) that mediate interactions with eukaryotic transcription factors. The p53 TAD interacts with CBP/p300 at multiple sites, and binding to one or more of the TAZ1, TAZ2, NCBD and KIX domains is required for CBP/p300-mediated transcription (14–17, 27–30). Deletion of either the AD1 or AD2 subdomains or mutation of key hydrophobic residues abrogates binding (14, 27). Because p53 binds DNA as a tetramer, 4 independent copies of the activation domain (one from each p53 subunit) will be presented at a promoter and multivalent interactions with the potential binding domains on a single CBP or p300 coactivator molecule are likely (31).
The role of the AD1 and AD2 subdomains in mediating binding of the p53 TAD to CBP is poorly understood. In the present article, we report a quantitative analysis of the relative binding affinities of the TAD and the AD1 and AD2 regions for the various CBP interaction domains. We show that binding to CBP is dominated by the AD2 subdomain, that AD1 makes the primary interactions with HDM2, and that the p53 TAD is capable of forming ternary complexes in which it is bound simultaneously to both CBP and HDM2. Phosphorylation at T18 impairs the interaction with HDM2 and slightly enhances binding to the KIX domain of CBP. Multisite phosphorylation (at S15, T18, and S20) greatly enhances binding to the TAZ1 domain, suggesting a mechanistic basis by which posttranslational modification releases HDM2, stabilizes p53, and promotes its transcriptional activity.
Results
The full-length p53 TAD spans residues 1–63 (21), but a construct containing only residues 13–61 was just as efficient as a longer construct (residues 1–94) in pulling down the TAZ1, TAZ2, KIX, and NCBD domains of CBP, and p53 (13–61) and p53 (1–61) exhibited similar binding affinities for NCBD and KIX, as measured by ITC. Truncated TAD constructs were therefore used for many experiments. To probe interactions with the isolated AD1 motif, 2 peptides, p53 (14–28) and p53 (13–37), which have comparable binding affinities for the CBP domains, were used. The interactions of the isolated AD2 motif were probed using peptide p53 (38–61).
Affinity of the p53 TAD for CBP Domains and HDM2.
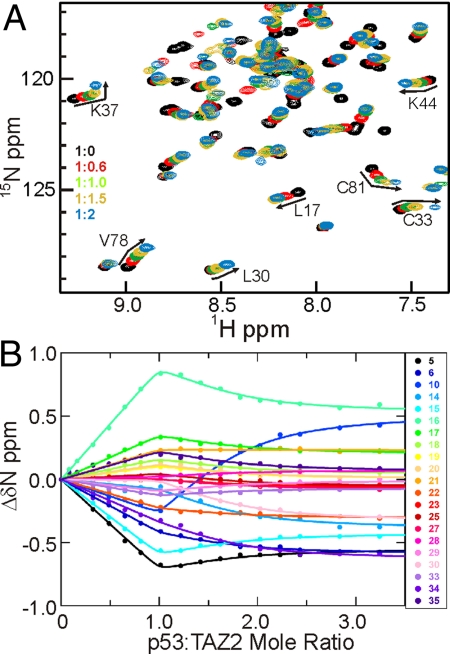
Affinities of the p53 TAD peptides for the CBP domains and the N-terminal domain of HDM2 were determined by ITC or from chemical shift changes in HSQC spectra of the CBP domains upon titration with p53 peptides (Table 1). Normally it is not possible to measure dissociation constants smaller than a few μM from chemical shift titrations; however, as we show in Fig. S1, under conditions of 2-site binding it is possible to accurately determine Kd values in the nM range. Representative HSQC titration data are shown in Fig. 2. (Additional data are shown in Figs. S2–S5.) The highest affinity for unphosphorylated p53 (13–61) is displayed by the TAZ2 domain, which binds 10 times more tightly than HDM2. The TAZ1 and NCBD domains bind ≈30- and 100-fold more weakly than TAZ2, and the KIX domain binds 1000-fold more weakly. Weak secondary binding sites for the p53 peptides are displayed by TAZ1 [Kd 310 μM for p53 (13–61)] and TAZ2 (Kd 30 μM), as evidenced by curvature in the chemical shift titration curves at p53 concentrations beyond 1:1 stoichiometry. It is worth noting that the high affinity Kd determined by NMR (26 nM and 0.9 μM for TAZ2 and TAZ1, respectively) are in excellent agreement with values measured by fluorescence anisotropy (27 nM and 1.1 μM) for binding of p53 (1–57) to the corresponding domains of p300 (31). Affinities of the CBP domains and HDM2 for shorter peptides representing the isolated AD1 and AD2 motifs (Table 1) indicate that the AD2 peptide binds with much higher affinity to the TAZ1, TAZ2, and NCBD domains than peptides containing only AD1. Nevertheless, AD1 does contribute to the overall interactions made by the full-length p53 TAD, because p53 (13–61) binds with significantly higher affinity than p53 (38–61), which contains only the AD2 motif. Binding of p53 (14–28) and p53 (38–61) to KIX is not detectable using ITC, although weak interactions can be observed by NMR (Fig. S5). HDM2 binds with high affinity to p53 (14–28), which contains the AD1 motif, and shows no detectable binding to the AD2 peptide by ITC.
Table 1.
Dissociation constants (Kd, μM) for the interactions of the p53 transactivation domain with CBP domains and HDM2
| Method | NMR |
ITC |
|||||
|---|---|---|---|---|---|---|---|
| CBP TAZ2 |
CBP TAZ1 |
CBP NCBD | CBP KIX | HDM2 | |||
| 1st site | 2nd site | 1st site | 2nd site | ||||
| p53(1–61) | nm | nm | nm | nm | 1.7 ± 0.3 | 19 ± 5 | 0.26 ± 0.02 |
| p53(13–61) | 0.026 ± 0.007* | 30 ± 3 | 0.9 ± 0.2 | 310 ± 40 | 3.1 ± 0.2 | 22 ± 5 | 0.23 ± 0.02 |
| p53(13–57) pT18 | 0.05 ± 0.02 | 56 ± 9 | 0.5 ± 0.1 | 330 ± 70 | 3.6 ± 0.4 | 5.2 ± 0.2 | 5 ± 1 |
| p53(13–57) pS15pT18pS20 | 0.08 ± 0.03 | 36 ± 10 | 0.07 ± 0.04 | 110 ± 25 | nm | 2.5 ± 0.3 | nm |
| AD1 peptide Kd | p53(13–37) 27 ± 2 | p53(13–37)177 ± 6 | p53(13–37) 350 ± 20 | p53(13–37) N/O† | p53(14–28) >100 | p53(14–28) weak‡ | p53(14–28) 0.90 ± 0.02 |
| p53(14–28) pT18 | nm | nm | nm | nm | 40 ± 20 | > 100 | 15 ± 3 |
| AD2 peptide p53(38–61) | 0.055 ± 0.006 | 10.1 ± 0.3 | 4.9 ± 0.7 | 192 ± 8 | 13.5 ± 0.5 | weak‡ | nd |
nm, not measured; N/O, not observed; nd, binding not detectable by ITC.
*Fitting error.
†One-site binding model fits the data, probably because the secondary binding is very weak.
‡Weak binding is observed by NMR but is not detectable by ITC.
Fig. 2.
Addition of p53 TAD to TAZ2. (A) Portion of the 1H-15N HSQC spectrum of TAZ2 (black) showing chemical shift changes upon titration with p53 (13–61) at p53:TAZ2 mole ratios of 0.6:1 (red), 1:1 (green), 1.5:1 (yellow), and 2:1 (blue). The curvature in the titrations with excess p53 indicates the presence of a secondary binding site. (B) 15N chemical shift titration curves for a subset of TAZ2 resonances (colored points corresponding to residues according to the legend) upon titration with increasing amounts of p53 (13–61). The lines represent a global fit to the titration data, using a 2-site binding model with Kd1 = 0.026 μM and Kd2 = 30 μM.
Effects of Phosphorylation on p53 Interactions with CBP and HDM2.
Phosphorylation at T18, in the peptide p53 (13–57)pT18, causes a 2-fold decrease in affinity for the TAZ2 domain compared with unphosphorylated p53 (13–61), no change in affinity for the NCBD, and a 2–4-fold increase in affinity for binding to TAZ1 and KIX (Table 1). In contrast, T18 phosphorylation results in a 20-fold decrease in affinity for HDM2; p53 (13–57)pT18 therefore binds to the TAZ1 domain with ≈10-fold higher affinity than it does HDM2. The affinity for all CBP domains is very much weaker in the absence of the AD2 motif, as evidenced by the Kd values for the complexes of p53 (14–28)pT18 with KIX and NCBD (Table 1) and by ITC profiles for binding to TAZ1 and TAZ2. Thus, as with the unphosphorylated TAD, both AD1 and AD2 contribute to binding.
The effects of multisite phosphorylation in the AD1 motif were investigated using a synthetic peptide phosphorylated at S15, T18, and S20. The binding affinity of p53 (13–57)pS15pT18pS20 for KIX was measured by ITC and for the TAZ1 and TAZ2 domains by HSQC titrations (Table 1). Triple phosphorylation causes no change in affinity for TAZ2 but strengthens binding to KIX and TAZ1 ≈10-fold. In marked contrast to the unphosphorylated TAD, the triply phosphorylated p53 peptide binds with the same affinity (≈70 nM) to both the TAZ1 and TAZ2 domains.
NMR Detection of p53 Interactions with CBP Domains and HDM2.
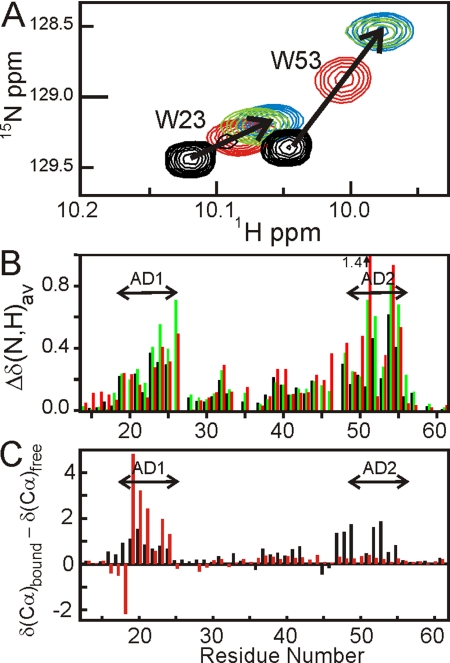
HSQC titrations were used to identify the sites at which the CBP domains and HDM2 (6–125) bind to the p53 TAD. Consistent with previous studies (23, 32), the 1H resonances of p53 (13–61) (Fig. S6) are poorly dispersed, showing that the p53 TAD is largely unfolded in the absence of a binding partner. Addition of each of the CBP domains results in extensive shifts of the p53 cross peaks (Fig. S7), indicating that p53 (13–61) changes conformation as a result of binding. As an illustration, the shifts of the tryptophan Nε cross peaks of p53 (13–61) upon titration with the NCBD construct are shown in Fig. 3A. Large changes are observed in the backbone amide 1H and/or 15N chemical shifts for residues 19–26 (AD1) and 48–55 (AD2) upon binding to the TAZ1, NCBD, and KIX domains (Fig. 3B), and backbone resonances from these same regions are also shifted and/or broadened upon binding to TAZ2 (Fig. S7). Smaller chemical shift changes are observed for residues 28–44 of the p53 TAD upon binding to each of the CBP domains, suggesting that regions between the AD1 and AD2 motifs also contact CBP or undergo a conformational change upon complex formation. Changes in 13Cα chemical shifts of p53 (13–61) upon binding to the NCBD (Fig. 3C, black bars) show that the interaction leads to an increase in helical structure in both the AD1 and AD2 regions, confirming that both motifs participate in the interactions between the p53 TAD and each of the CBP domains. In contrast, HDM2 (6–125) binds to the p53 TAD predominantly through the AD1 motif, leading to stabilization of helical structure for residues 19–24, although very weak interactions are also evident for AD2 (red bars in Fig. 3C and ref. 33).
Fig. 3.
Addition of CBP domains to p53 TAD. (A) Chemical shift changes for Trp-23 and Trp-53 Nε cross peaks upon titration of 15N p53-TAD (13–61) with unlabeled NCBD. Mole ratio p53:NCBD = 1:0 (black), 1:0.5 (red), 1:1 (green), 1:2 (blue). (B) Histogram showing weighted average chemical shift changes Δδ(N,H)av (= , where ΔδHN and ΔδN correspond to the differences in amide 1H and 15N chemical shifts between the free and bound states) for p53 amide resonances caused by binding to TAZ1 (black), KIX (green) and NCBD (red). (C) Histogram showing changes in 13Cα chemical shifts upon binding of 15N,13C p53-TAD (13–61) to NCBD (black) or to HDM2 (6–125) (red).
Ternary Complex Formation by CBP, p53, and HDM2.
Because the AD1 and AD2 motifs of the p53 TAD bind preferentially to HDM2 and to the CBP domains, respectively, we performed NMR titrations to determine whether the p53 TAD can form a ternary complex, with one of the CBP domains bound to the AD2 motif and with HDM2 bound to AD1. Fig. 4A shows part of the HSQC spectrum of free 15N-labeled p53 (13–61). Addition of equimolar HDM2 to the p53 TAD results in large shifts and significantly greater dispersion of the cross peaks of residues in the AD1 motif (red arrows in Fig. 4A) but causes little or no change in chemical shifts of residues in the AD2 motif. Subsequent addition of equimolar TAZ2 does not affect the AD1 resonances, which remain in the same positions as for the binary HDM2 complex, but resonances of AD2 residues are shifted or broadened, e.g., the cross-peak of Thr-55 is strongly shifted on binding of TAZ2 (green arrow in Fig. 4A). Addition of equimolar HDM2 to the binary complex formed from 15N-labeled TAZ2 and p53 (13–61) results in shifts of some HSQC cross-peaks of TAZ2, specifically for residues that form the binding site for AD1. These results show that HDM2 competes with TAZ2 for binding AD1 but has no effect on the interactions of TAZ2 with the AD2 motif.
Fig. 4.
Ternary complex formation between CBP, p53, and HDM2. (A) HSQC spectra of 15N p53 (13–61) free (black), bound to equimolar HDM2 (red), and in 1:1:1 ternary complex with TAZ2 and HDM2 (green). (B) Tryptophan side chain resonances of 15N p53 (13–61) free (black), bound to equimolar HDM2 (red), and after addition of equimolar TAZ1 to the HDM2 complex (green). For reference, the positions of the Trp Nε cross peaks in the binary p53 (13–61):TAZ1 complex are shown in blue.
Formation of a ternary complex is illustrated by the chemical shift changes of the tryptophan Nε resonances (Fig. 4B). Addition of HDM2 to 15N-labeled p53 (13–61) causes a large shift in the Nε resonance of Trp-23 (in AD1) but very little shift in the cross peak of Trp-53 (in AD2). Subsequent addition of equimolar TAZ1 causes the Nε cross peak of Trp-53 to shift significantly, confirming binding of TAZ1 to AD2, whereas the Trp-23 cross peak remains at precisely the same chemical shifts as in the binary p53:HDM2 complex. These data show conclusively that both HDM2 and TAZ1 remain bound to the p53 TAD, through the AD1 and AD2 motifs, respectively. If the TAZ domains were to compete with HDM2 for binding to p53, then addition of TAZ1 or TAZ2 to the binary p53:HDM2 complex would either yield the spectrum of the binary p53:TAZ1/2 complex (blue contours for TAZ1 in Fig. 4B) or, if a mixture of binary HDM2 and TAZ complexes was formed, give rise to a spectrum that is a superposition of the spectra of the component binary complexes. Neither of these scenarios is observed and the spectra of the 1:1:1 p53:HDM2:TAZ1/2 mixture are unique and show definitively that the AD1 and AD2 motifs are fully bound to HDM2 and TAZ1/2, respectively, within a ternary complex (Fig. 4B and Fig. S8). Direct confirmation that a ternary complex is formed between the p53 TAD, HDM2 and the TAZ1 domain was obtained by pulldown experiments (SI Text and Fig. S9). Our results are inconsistent with published binding mechanisms (31) in which HDM2 and the TAZ domains compete for binding to the p53 TAD. HSQC titrations show that the NCBD and KIX domains also form ternary complexes with p53 and HDM2 (Fig. S8).
The changes in the Trp-53 Nε chemical shifts upon titration of TAZ2 into 15N-labeled p53 (38–61) (AD2 motif) to form a binary complex, and into the 15N-p53 (13–61):HDM2 complex to form the ternary complex allowed an estimate of the binding affinity (Fig. S9). For both the binary and ternary complexes, the Trp-53 chemical shift changes fit to a 2-site binding model with the same dissociation constants (Kd1 = 0.055 μM, Kd2 = 10.1 μM) determined from global fitting of the HSQC titration of p53 (38–61) into 15N-labeled TAZ2 (Table 1). Thus, the presence of HDM2 bound at the AD1 site in the full-length p53 TAD neither strengthens nor weakens subsequent interactions between the AD2 motif and TAZ2; the 2 motifs function independently in forming the ternary complex. Further, there appear to be no interactions between HDM2 and the TAZ domains within the ternary complex; the AD1 resonances have identical chemical shifts in the binary HDM2 complex and the ternary HDM2:p53:TAZ2 complex; similarly, the AD2 resonances in the binary TAZ2 and ternary HDM2:p53:TAZ2 complexes are the same. Neither TAZ1 (34) nor TAZ2 binds directly to the HDM2 N-terminal domain: When 15N-labeled TAZ2 is mixed with HDM2 (6–125) in the absence of p53, there is no change in the HSQC spectrum of TAZ2. Upon formation of the ternary complex, the HSQC cross-peaks of many residues located between the AD1 and AD2 motifs of p53 (13–61) remain intense and are shifted only slightly from their positions in the free p53 spectrum, suggesting that this region remains disordered and does not interact significantly with the bound HDM2 or TAZ2 domains.
Further evidence that the TAZ2:AD2 and HDM2:AD1 domains tumble independently comes from the fact that linewidths of cross-peaks in the HSQC spectrum of the 15N-TAZ2:p53 (13–61) complex do not increase upon binding of HDM2 to form the ternary complex, despite the overall increase in molecular weight (see for example Fig. 4B). This behavior is as expected for a hydrodynamic model in which the TAZ2:AD2 and HDM2:AD1 domains tumble as independent beads on a flexible string. Thus, in the ternary complex, the AD1 and AD2 regions function as independent binding motifs, separated by a flexible linker, that can interact simultaneously with HDM2 and CBP domains (Fig. 5A).
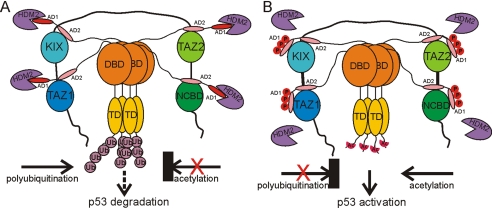
Fig. 5.
Model for the molecular events involved in the p53 response mediated by synergistic interactions with CBP/p300 and HDM2. (A) In unstressed cells, AD1 binds strongly to HDM2, whereas AD2 interacts with the TAZ1, KIX, TAZ2, and NCBD domains of CBP/p300, promoting polyubiquitination and degradation of p53. (B) After genotoxic stress, S15, T18, and S20 of the p53 TAD become phosphorylated, lowering the affinity of AD1 for HDM2 and increasing its affinity for the CBP TAZ1 and KIX domains, promoting acetylation of the C terminus of p53 and its activation and stabilization.
Discussion
Our results provide new insights, at the molecular level, into the complex interplay between CBP/p300, p53, and HDM2. They are fully consistent with previous reports (14–16, 28–30) of binding of the p53 N-terminal TAD at multiple sites on CBP/p300, but go beyond these to provide a quantitative measure of the relative binding affinities for each site and to investigate the effects of multisite phosphorylation. The unphosphorylated transactivation domain (p53 residues 13–61, encompassing both the AD1 and AD2 regions) binds with highest affinity to the TAZ2 domain of CBP, and with intermediate affinity to the TAZ1 and NCBD domains. The interaction with KIX is significantly weaker. Our affinity measurements are consistent with recent fluorescence anisotropy studies of binding of p53 (1–57) and p53 (15–29) peptides to the corresponding domains from p300 (31), with some small differences in binding affinities probably reflecting differences in temperature or slight sequence differences between the CBP and p300 domains. Because all 4 CBP domains bind the p53 TAD with moderate to high affinity, it is likely that each of them contributes to high affinity binding of the p53 tetramer, as suggested recently for p300 (31). Multivalent interactions of p53 with the STAGA complex have also been reported (35).
Both the AD1 and AD2 regions of the p53 transactivation domain participate in the interactions with CBP, but with significantly different affinities. Peptides containing the isolated AD2 motif bind 70–500 times more tightly to the TAZ1 and TAZ2 domains than do those containing the AD1 motif alone. Although AD2 dominates the interaction, AD1 nevertheless contributes slightly (2- and 5-fold for TAZ2 and TAZ1, respectively) to the binding affinity of the full-length p53 TAD (Table 1) and is required for fully functional interactions between CBP and p53 in vivo (36). Chemical shift changes for AD1 resonances that accompany binding of p53 (13–61) suggest that the enhanced binding comes from direct contacts with the TAZ domains that lead to partial folding of the AD1 motif. This interpretation is supported by chemical shift changes observed in TAZ1 and TAZ2 resonances, which indicate a contact site on each TAZ domain for the AD1 residues. Similar binding preferences are observed for the NCBD, with the AD2 peptide binding ≈10-fold more tightly than AD1 whereas both motifs contribute to binding of the full-length p53 TAD. In the case of the KIX domain, both AD1 and AD2 are essential for high affinity binding; no binding of the isolated AD1 and AD2 peptides is detectable by ITC, although HSQC titrations indicate weak interactions.
Our experiments show that all 4 CBP domains, even KIX, form a ternary complex with AD1 bound to HDM2 and AD2 interacting with CBP (Fig. 4 and Fig. S8). Despite the high affinity with which TAZ2 binds to the p53 TAD, it is unable to displace the much more weakly bound HDM2, showing clearly that the AD1 and AD2 motifs function independently in their interactions with CBP and HDM2. This is also confirmed by the observation that the chemical shifts of AD1 resonances are the same in the binary complex with HDM2 and in the ternary complexes with the various CBP domains, and conversely for AD2 resonances in the presence or absence of HDM2. In addition, the binding affinity of the CBP domains for AD2 does not appear to be altered significantly by the presence of HDM2 in a ternary complex (Fig. S9). The full-length p53 TAD binds both the TAZ1 and NCBD domains more weakly (Kd ≈ 1 μM) than it binds HDM2, but the AD1 and AD2 motifs once again function independently: There is no competition between HDM2 and the CBP domains for binding to p53. Although independent binding of the AD2 motif to the individual TAZ1, NCBD, and KIX domains of CBP is relatively weak (Kd > 5 μM), simultaneous and synergistic binding of all 4 CBP domains to the 4 transactivation domains presented by the p53 tetramer would lead to a high affinity complex (Fig. 5A). A ternary complex between tetrameric p53, HDM2, and p300 has been isolated from HeLa nuclear extracts (37).
Although our NMR experiments (Fig. S8) and pulldown assays (Fig. S9) establish unequivocally that isolated CBP domains can bind the p53 TAD in the presence of bound HDM2 to form ternary complexes, there are contrary reports in the literature. A recent systematic study of the p53-binding properties of the p300 domains, using fluorescence anisotropy to monitor binding concluded that HDM2 and the CH3 domain (TAZ2 plus the ZZ domain) compete for binding to the p53 TAD (31).
We suspect that the reason Teufel et al. (31) failed to detect the ternary complex is because, as indicated by our NMR experiments, it behaves hydrodynamically as 2 beads on a flexible string and its formation is therefore not accompanied by an increase in rotational correlation time of the fluorescence probe. It has also been reported that HDM2 inhibits the interaction in vivo between p53 and constructs of CBP/p300 containing only the TAZ1 or TAZ2 domains (30). However, in vitro assays showed that HDM2 does not inhibit binding of p53 to full length p300 (38), suggesting that the ability of CBP/p300 to bind the p53 tetramer through multivalent interactions involving the TAZ1, TAZ2, NCBD, and KIX domains substantially enhances the binding affinity.
The p53 response is regulated by a cascade of phosphorylation events activated by various forms of genotoxic stress (4, 6). After DNA damage, the cascade is initiated by phosphorylation of S15 in the p53 TAD by ATM family kinases, with subsequent phosphorylation at T18 and S20 (13). In accord with the studies in refs. 8 and 9, we find that T18 phosphorylation significantly impairs HDM2 binding (Table 1). However, we see no evidence for significant enhancement of binding to CBP/p300, beyond a very small increase in binding affinity for the KIX domain. Because there is growing evidence that simultaneous phosphorylation of S15 and S20, or T18 and S20 has a synergistic role in activating p53-mediated apoptosis (39–41), we investigated binding of a p53 peptide phosphorylated at S15, T18, and S20 to the CBP domains. In contrast to the modest effect of single site phosphorylation at T18, triple site phosphorylation dramatically and specifically enhanced binding to TAZ1, increasing the affinity >10-fold such that the triply phosphorylated TAD binds with equal avidity (Kd ≈ 70 nM) to both the TAZ1 and TAZ2 domains. As noted in ref. 9, binding of a triply phosphorylated peptide, p53 (15–29)pS15pT18pS20, to MDM2 is impaired to the same extent as caused by phosphorylation at T18 alone.
CBP/p300 and HDM2 play a synergistic role in regulation of p53 stability. In unstressed cells, CBP/p300, p53 and HDM2 form a ternary complex that mediates p53 turnover (17). Binding to HDM2 alone promotes monoubiquitination of p53; interactions with the CBP/p300 N-terminal region, which functions as a ubiquitin E4 ligase, are required for polyubiquitination and proteasomal degradation of mono-ubiquitinated p53 (18). It was originally suggested that direct interactions between HDM2 and the TAZ1 domain were required for p53 degradation (17), but subsequent studies indicated that the observed interactions were an artifactual consequence of denaturation of the TAZ1 domain by sequestration of zinc (34). CBP and p300 also play an important role in stabilization of p53 by acetylation of lysine residues in the C-terminal regulatory region (7); acetylation of p53 is inhibited in the ternary complex formed with HDM2 (37).
Our data suggest a plausible model for the molecular events involved in the p53 response (Fig. 5). In the absence of genotoxic stress, the p53:HDM2 complex interacts relatively weakly with CBP/p300 leading to polyubiquitination and p53 turnover. In response to DNA damage, p53 becomes stabilized and activated by phosphorylation at multiple sites, leading to release of HDM2, enhanced recruitment of CBP/p300, and acetylation of the C-terminal regulatory domain of p53 (4–6). Phosphorylation at T18 inhibits binding of the p53 TAD to HDM2 (Kd increases to 5 μM) but has little effect on binding CBP/p300. In contrast, triple phosphorylation at S15, T18, and S20 enhances the affinity of the p53 TAD for the TAZ1 and KIX domains 10-fold (Table 1), while simultaneously impairing binding to HDM2 (9). This dramatic switch in binding affinity would shut off the polyubiquitination activity by inhibiting binding of the complex formed by HDM2 and the nonphosphorylated p53 TAD to TAZ1. This inhibitory function becomes especially important in the context of a DNA-bound p53 tetramer, where the 4 TAD regions, 1 from each p53 subunit, may be phosphorylated differently. By enhancing the affinity for binding to TAZ1, phosphorylation of S15, T18, and S20 in only 1 of the 4 TADs present in the tetramer would be sufficient to shut down ubiquitination, even if the other TADs are not phosphorylated and remain bound to HDM2.
Materials and Methods
Protein Expression and Purification.
Unlabeled and 15N-labeled TAZ1 (residues 340–439), KIX (residues 586–672), TAZ2 (residues 1764–1855), and NCBD (residues 2059–2117) domains of mouse CBP were expressed and purified as described (42–45). The p53 binding domain of HDM2, residues 6–125, was expressed in E. coli. Details of the purification of HDM2 and the preparation and purification of isotopically-labeled p53 constructs are given in SI Text.
Peptide Synthesis.
The peptide p53 (14–28) and the phosphorylated peptides p53 (14–28)pT18, p53 (13–57)pT18, and p53 (13–57)pS15pT18pS20 were synthesized on a Perseptive Biosystems synthesizer using solid-phase Fmoc methods with double coupling at each step. Fmoc-Thr(PO(OBzl)-OH)-OH and Fmoc-Ser(PO(OBzl)-OH)-OH were used for incorporation of phosphothreonine and phosphoserine, respectively. The peptides were purified by preparative reversed phase HPLC on a C18 silica column and purity and mass were confirmed by analytical reversed phase HPLC and MALDI-TOF.
Isothermal Titration Calorimetry.
Affinities of the p53 TAD constructs for the NCBD and KIX domains of CBP and for HDM2 were measured at 35 °C by isothermal titration calorimetry. Details are provided in SI Text and typical results shown in Fig. S2.
NMR Spectroscopy.
1H-15N HSQC titrations were performed to characterize binding of CBP domains to the p53 TAD constructs. Details of the NMR experiments and titration conditions are provided in SI Text.
Determination of Kd from NMR Titrations.
Accurate affinities for the TAZ1 and TAZ2 complexes were difficult to determine by ITC because of a weak secondary p53 binding site, indicated by curvature in HSQC titration curves at p53:TAZ ratios >1:1 (Fig. 2). Dissociation constants were therefore determined from changes in amide 1H (ΔδH) and 15N (ΔδN) chemical shifts. The ΔδH and ΔδN titration curves were fitted globally to 1-site or 2-site binding models with an in-house fitting program nmrKd, using the Levenberg–Marquardt algorithm (46). The 1-site binding model assumes
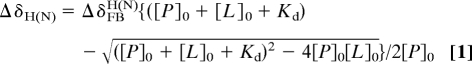 |
where ΔδFBH(N) is the amide proton (nitrogen) chemical shift difference between the free and bound form, and [P]0 and [L]0 are the total concentrations of CBP domain and p53 peptide, respectively (47, 48). The 2-site binding model assumes
where Kd1, ΔδFB1H(N) and Kd2, ΔδFB2H(N) are Kd and ΔδFBH(N) for primary and secondary binding, respectively, and [L] is the free p53 concentration, which is obtained analytically in terms of Kd1, Kd2, [P]0, and [L]0 as described in ref. 49. Details of the fitting procedure will be described elsewhere.
Because TAZ1 and TAZ2 concentrations could not be determined accurately by UV absorbance because of the presence of DTT, concentration corrections were implemented in the fitting program. Thus, [P]0 was replaced by c[P]0 in the above equations, where c is a correction factor for TAZ1 or TAZ2 concentration and is set as a variable in the fitting. The correction led to improved fits. The c value was typically 0.7 and 1.2 for TAZ1 and TAZ2, respectively.
Supplementary Material
Acknowledgments.
We thank John Chung and Gerard Kroon for expert NMR assistance and Linda Tennant, Mindy Landes and Euvel Manlapaz for technical support. This work was supported by National Institutes of Health Grant CA96865, the Skaggs Institute for Chemical Biology, Korea Research Foundation Grant KRF-2004-214-C00207 funded by the Korean Ministry of Education and Human Resource Development, Basic Research Promotion Fund (to C.W.L.), and a Skaggs training grant (to C.W.L.). J.C.F is a Leukemia and Lymphoma Society Special Fellow.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811023106/DCSupplemental.
References
- 1.Espinosa JM, Verdun RE, Emerson BM. p53 Functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 2.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 5.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi K, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 9.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 10.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 12.Dornan D, Hupp TR. Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300. EMBO Rep. 2001;2:139–144. doi: 10.1093/embo-reports/kve025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito S, et al. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem. 2003;278:37536–37544. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 14.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 15.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 16.Avantaggiati ML, et al. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 17.Grossman SR, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 18.Grossman SR, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 19.Unger T, Mietz JA, Scheffner M, Yee CL, Howley PM. Functional domains of wild-type and mutant p53 proteins involved in transcriptional regulation, transdominant inhibition, and transformation suppression. Mol Cell Biol. 1993;13:5186–5194. doi: 10.1128/mcb.13.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Candau R, et al. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Zhang S, Jiang J, Chen X. Definition of the p53 functional domains necessary for inducing apoptosis. J Biol Chem. 2000;275:39927–39934. doi: 10.1074/jbc.M005676200. [DOI] [PubMed] [Google Scholar]
- 22.Ayed A, et al. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 23.Dawson R, et al. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 25.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Scolnick DM, et al. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 28.Livengood JA, et al. p53 transcriptional activity Is mediated through the SRC1-interacting domain of CBP/p300. J Biol Chem. 2002;277:9054–9061. doi: 10.1074/jbc.M108870200. [DOI] [PubMed] [Google Scholar]
- 29.van Orden K, Giebler HA, Lemasson I, Gonzales M, Nyborg JK. Binding of p53 to the KIX domain of CREB binding protein - A potential link to human T-cell leukemia virus, type I-associated leukemogenesis. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 30.Wadgaonkar R, Collins T. Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J Biol Chem. 1999;274:13760–13767. doi: 10.1074/jbc.274.20.13760. [DOI] [PubMed] [Google Scholar]
- 31.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H, et al. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 33.Chi SW, et al. Structural details on mdm2–p53 interaction. J Biol Chem. 2005;280:38795–38802. doi: 10.1074/jbc.M508578200. [DOI] [PubMed] [Google Scholar]
- 34.Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. The CBP/p300 TAZ1 domain in its native state is not a binding partner of MDM2. Biochem J. 2004;381:685–691. doi: 10.1042/BJ20040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Mol Cell Biol. 2008;28:2517–2527. doi: 10.1128/MCB.01461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao C, et al. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobet E, Zeng XY, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci USA. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jabbur JR, Zhang W. p53 Antiproliferative function is enhanced by aspartate substitution at threonine 18 and serine 20. Cancer Biol Ther. 2002;1:277–283. doi: 10.4161/cbt.81. [DOI] [PubMed] [Google Scholar]
- 40.Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–2622. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamizo A, et al. Phosphorylation of Thr18 and Ser20 of p53 in Ad-p53-induced apoptosis. Neuro-oncol. 2008;10:275–291. doi: 10.1215/15228517-2008-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 43.De Guzman RN, Liu HY, Martinez-Yamout M, Dyson HJ, Wright PE. Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP. J Mol Biol. 2000;303:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- 44.Demarest SJ, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 45.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Press, et al. Numerical Recipes in C. Cambridge, UK: Cambridge Univ Press; 1992. [Google Scholar]
- 47.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Prog NMR Spectros. 2007;51:219–242. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 48.Cavanagh , et al. Protein NMR Spectroscopy: Principles and Practice. Burlington, MA: Elsevier Academic; 2007. [Google Scholar]
- 49.Wang ZX, Jiang RF. A novel two-site binding equation presented in terms of the total ligand concentration. FEBS Lett. 1996;392:245–249. doi: 10.1016/0014-5793(96)00818-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.