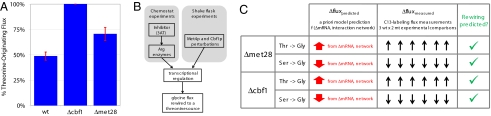
Fig. 5.
Observations in glycine flux rewiring demonstrate a biological principle and validate the predictive network model. (A) Additional transcriptional regulatory perturbations drive a rewiring in glycine flux. Knockout strains for transcriptional regulators in the vicinity of glycine biosynthesis (met28Δ, cbf1Δ, met31Δ, and met32Δ) were constructed, mRNA levels were measured, and biosynthetic fluxes were determined for the 2 strains (met28Δ and cbf1Δ) that displayed significant transcriptional changes. The results for the percentage of threonine-originating glycine flux are displayed in the first panel. Similar to the previous Gcn4p-induced stress response data, we observe a rewiring of glycine flux away from serine precursor in both the met28Δ and cbf1Δ cultures. (B) Convergent rewiring of glycine flux. In both chemostat and shake flask experiments, transcriptional regulatory networks drives a rewiring of glycine flux to a threonine source. (C) The predictive model from Gcn4 experiments uses mRNA changes and the interaction network correctly identified flux rewiring in knockout experiments. mRNA changes from the met28Δ and cbf1Δ knockout strains were input to the existing predictive flux model to assess glycine flux rewiring. For both knockout mRNA profiles, the glycine flux was predicted to be rewired from a serine precursor to threonine. The experimentally measured flux changes of the serine and glycine (displayed in A) verify the prediction of this observed rewiring.