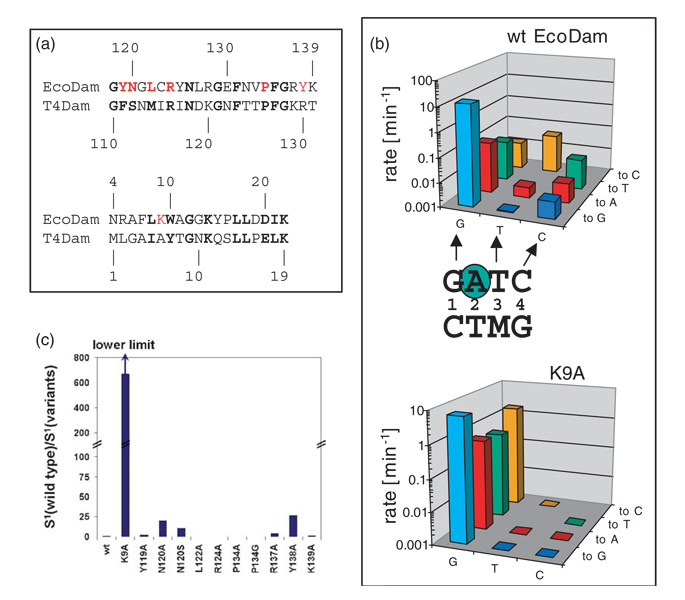
Figure 3. Recognition of the first base-pair by N-terminal K9.
(a) Pair-wise sequence alignment of EcoDam and T4Dam in two regions: the β hairpin loop and the N-terminal loop. The residues colored in red were targets for site-directed mutagenesis. (b) Specificity profile of EcoDam wild-type (top panel) and the K9A variant (bottom panel). For easy comparison, we duplicate the wild-type panel.20 The single-turn-over methylation rates of the wild-type and the K9A variant are given for the cognate, hemimethylated GATC substrate (light blue bars) as well as for all nine near-cognate hemimethylated substrates. On the horizontal axis, the three positions of the GATC site that are mutated are given (G=GATC, T=GATC, C=GATC, M=N6 mA). The new base introduced at each position is specified on the right-hand axis. The methylation rates of the respective pair of enzyme and substrate are given on the vertical axis (note the logarithmic scale). (c) Specificity factor (defined in Materials and Methods) of EcoDam variants for recognition of the first position of the GATC sequence (S1). The values are given as relative changes with respect to the wild-type. Because no activity could be detected at near-cognate sites modified at the third or fourth base-pair of GATC with the K9A variant, the S1 factor given here is a lower limit, indicated by the arrow. The specificity factors of wild-type EcoDam and the K9A were calculated using the data given in (b) and (c); the data for all other variants were taken from Horton et al.20