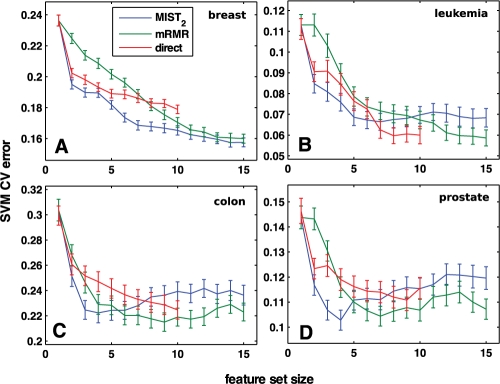
Fig. 4.
Gene subset selection for cancer classification. Subsets of gene expression levels were chosen incrementally to maximize the information with the cancer class according to MIST2, direct estimation of MI or mRMR and scored by the cross-validation error of an SVM classifier. For all data sets, 75% of the data were separated and used to select features and train the model; the classifier was then used to classify the remaining 25%. The mean classification error and standard error of the mean for 200 training/testing partitionings are reported. Genes were selected for data sets relating to (A) breast, (B) leukemia, (C) colon and (D) prostate cancer.