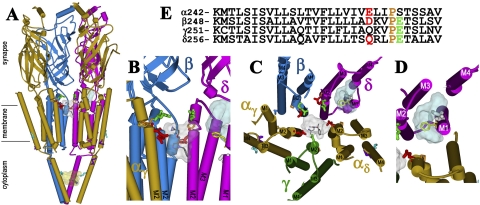
Fig. 8.
The binding sites for [3H]azietomidate in the nAChR. Views of the T. marmorata nAChR structure (Protein Data Bank code 2BG9) (α, gold; β, blue; γ, green; δ, magenta) showing α-helical (cylinders) and β-sheet (ribbon) secondary structure. A, a perspective parallel to the membrane surface (with the γ subunit omitted). B, an expanded view of A focused on the top of the channel (γ and αδ omitted). C, the transmembrane helices viewed looking down the ion channel. D, the δ subunit helix bundle looking down the M1 helix. E, the amino acid sequences of each of the M2 helices of the nAChR structure, with the amino acids highlighted in the structures indicated by the same colors. The residues labeled by [3H]azietomidate are shown in stick format, color-coded for location: ion channel, position M2-20 (αGlu-262, βAsp-269; and δ-Gln-276; red); the δ subunit helix bundle (δCys-236, white); αM4 (αCys-412; cyan); the cytoplasmic basket formed by the MA helices (αGlu-390, cyan). Also indicated in stick format in the M2 helices are unlabeled acidic side chains (βGlu-273 and δGlu-280; green) that project into the channel lumen and the prolines (orange) that precede those positions in each subunit. δPhe-232 (yellow), the amino acid in δM1 that is photolabeled by [125I]TID (Arevalo et al., 2005) and [3H]benzophenone (Garcia et al., 2007), is included, as are the amino acids in αMA/αM4 (αGlu-398, αAsp-407, purple) photolabeled by [3H]azicholesterol (Hamouda et al., 2006). The volumes defined by the ensemble of the 10 lowest energy orientations of azietomidate docked at the extracellular end of the ion channel (gray, 680 Å3), in the δ subunit helix bundle (blue, 570 Å3), and in the cytoplasmic basket (yellow, 970 Å3) are shown in Connolly surface representations with a single azietomidate docked in its lowest energy orientation in each pocket (see Materials and Methods).