Table 1. Data-collection and refinement statistics.
Values in parentheses are for the outer shell.
| EfsQnr (modified) | Sm acetate | |
|---|---|---|
| Space group | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 36.9, b = 63.1, c = 94.6, β = 96.1 | a = 36.6, b = 65.5, c = 94.1, β = 97.5 |
| Resolution (Å) | 20–1.6 (1.69–1.6) | 38–2.0 (2.11–2.0) |
| Completeness (%) | 94.7 (86.0) | 100 (100) |
| Redundancy | 2.9 (2.7) | 7.5 (3.9) |
| Mean (I)/σ(I) | 19.3 (3.1) | 20.3 (4.1) |
| Rmerge† | 0.043 (0.253) | 0.091 (0.322) |
| Wilson B factor (Å2) | 22.4 | 15.4 |
| Model and refinement data | ||
| Resolution (Å) | 20–1.6 (1.64–1.6) | |
| Unique reflections | 51172 (3093) | |
| Rcryst‡ (%) | 18.7 (36.0) | |
| Rfree‡ (5% of data) (%) | 22.1 (44.9) | |
| Contents of model | ||
| Residues (1–211 + tag) | A1–A8, A13–A211, B4–B211 | |
| Waters | 417 | |
| Other | 4 Cl | |
| Total atoms | 3910 | |
| Average B factor (Å2) | ||
| Protein | 13.6 | |
| Waters | 29.7 | |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.014 | |
| Angles (°) | 1.59 | |
| MOLPROBITY statistics | ||
| Ramachandran favored/outliers (%) | 98.2/0.0 | |
| Rotamer outliers (%) | 1.1 | |
| Clashscore§ | 7.2 [85th percentile, N = 718, 1.35–1.85 Å] | |
| Overall score§ | 1.4 [92nd percentile, N = 7200, 1.35–1.85 Å] |
R
merge = 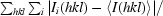
 , where I
i(hkl) is the intensity measurement for reflection i and 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
, where I
i(hkl) is the intensity measurement for reflection i and 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
R
work, R
free = 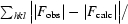
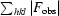 , where the working and free R factors are calculated using working and free reflections, respectively.
, where the working and free R factors are calculated using working and free reflections, respectively.
As calculated using the MOLPROBITY server; N is the number of sample structures in the resolution range.
