Abstract
Study Objectives
Sleep is regulated by circadian and homeostatic processes. Recent studies with mutant mice have indicated that circadian-related genes regulate sleep amount, as well as the timing of sleep. Thus a direct link between circadian and homeostatic regulation of sleep may exist, at least at the molecular level. Prokineticin 2 (PK2), which oscillates daily with high amplitude in the suprachiasmatic nuclei (SCN), has been postulated to be an SCN output molecule. In particular, mice lacking the PK2 gene (PK2−/−) have been shown to display significantly reduced rhythmicity for a variety of circadian physiological and behavioral parameters. We investigated the role of PK2 in sleep regulation.
Design
EEG/EMG sleep-wake patterns were recorded in PK2−/− mice and their wild-type littermate controls under baseline and challenged conditions.
Measurements and Results
PK2−/− mice exhibited reduced total sleep time under entrained light-dark and constant darkness conditions. The reduced sleep time in PK2−/− mice occurred predominantly during the light period and was entirely due to a decrease in non-rapid eye movement (NREM) sleep time. However, PK2−/− mice showed increased rapid eye movement (REM) sleep time in both light and dark periods. After sleep deprivation, compensatory rebound in NREM sleep, REM sleep, and EEG delta power was attenuated in PK2−/− mice. In addition, PK2−/− mice had an impaired response to sleep disturbance caused by cage change in the light phase.
Conclusions
These results indicate that PK2 plays roles in both circadian and homeostatic regulation of sleep. PK2 may also be involved in maintaining the awake state in the presence of behavioral challenges.
Keywords: Sleep, prokineticin 2, sleep homeostasis, circadian genes, EEG, behavioral challenge
INTRODUCTION
Homeostatic and circadian processes have been postulated to regulate sleep.1 The homeostatic process determines the duration and intensity of sleep, whereas the circadian process, driven by the suprachiasmatic nuclei (SCN) of the hypothalamus in mammals, determines the timing of sleep.2 Recently, studies of circadian gene mutant mice have revealed that circadian genes affect not only circadian sleep distribution, but also the homeostatic regulation of sleep. Clock gene mutant mice sleep 2 h less than wild-type mice daily,3 whereas Bmal1/Mop3 deficient mice and double Cry1 and Cry2 deficient mice display 1.5 h and 1.8 h increases in total daily sleep amount,4,5 respectively. In wild-type mice, sleep deprivation (SD) is followed by a compensatory sleep increase. Mice lacking both Cry1 and Cry2 did not exhibit rebound in non-rapid eye movement (NREM) and rapid eye movement (REM) sleep time,5 while mice lacking Clock or Bmal1/Mop3 have a reduced REM sleep rebound in response to SD.3,4 Mutation of the DBP gene, encoding a clock-controlled transcription factor, also leads to a decrease in REM sleep amount, in addition to a reduction in circadian amplitude of NREM sleep amount.6
Prokineticin 2 (PK2) is a postulated SCN output molecule that regulates circadian rhythm.7–9 PK2 oscillation in the SCN is regulated by CLOCK and BMAL1 binding to E-boxes on the PK2 promoter.7 PKR2, the G-protein coupled receptor activated by PK2, is expressed in the primary SCN output target regions, including the paraventricular and paratenial nuclei of the thalamus, the paraventricular nucleus of the hypothalamus, the dorsal medial nucleus of the hypothalamus, the lateral septal nucleus, and the bed nucleus of the stria terminalis.7,10 Recently, PK2-deficient (PK2−/−) mice have been shown to display significantly reduced rhythmicity for a variety of physiological and behavioral parameters, including locomotor activity, body temperature, food intake, circulating glucocorticoid and glucose levels, and the expression of peripheral clock genes.11 Thus, PK2 is critical for the maintenance of robust circadian rhythms, acting as an SCN output factor.
In this study, we investigated the sleep parameters of PK2−/− mice under both baseline and sleep deprived conditions, as well as sleep response to environmental challenges.
MATERIALS AND METHODS
Animals
PK2−/− mice and their littermate wild-type controls on a C57BL/6J : 129/Ola background were used for these experiments. The breeding and genotyping of PK2−/− mice were performed as described elsewhere.11 All mice were 11 to 20 weeks old and weighed 22–28 g. All procedures regarding the care and use of animals were in accordance with institutional guidelines.
Surgery Procedures
Mice were anesthetized with ketamine-xylazine (100 and 10 mg/kg, i.p.), and standard sterile surgical and stereotaxic procedures were employed for implant placements. The cranium was exposed, and 4 burr holes were drilled, anterior and posterior to bregma, bilaterally (AP 1.1, ML ± 1.45 and AP − 2.5, ML ± 1.45). Stainless steel screws were then inserted into these holes to record electro-encephalographic (EEG) signals. A pair of Teflon coated stainless steel wires with exposed ends were inserted into the dorsal neck muscles to record electromyogram (EMG) signals. Electrodes were cemented to the skull with dental acrylic. All electrodes were then connected to a plastic connector (Plastics One, Roanoke, VA) that was secured to the skull using dental acrylic. After surgery, animals were housed in individual cages (20 × 20 × 30 cm) with a 12 h : 12 h light : dark (LD) cycle (lights on at 07:00 and off at 19:00) and at a constant temperature (22–24°C). They had ad libitum access to food and water. They were allowed 10–14 days to recover, during which they were habituated to the recording conditions, with the home cage being the recording cage.
Sleep Recording
The mice were connected to a wire tether / commutator system (Plastics One, Roanoke,VA) for the collection of EEG and EMG signals. This swivel system allowed the animal unrestricted movement throughout the recording cage. After at least 5 days of adaptation to the recording environment, a 48-hour baseline EEG/EMG recording was collected on a LD cycle with lights on at 7:00 and off at 19:00. Mice were recorded concurrently in matched littermate pairs of PK2−/− and wild-type controls. EEG/EMG signals were amplified using a Grass Telefactor Model 15LT with 15A94 amplifier (Grass Instruments, West Warwick, RI) and filtered (EEG: 0.3–100 Hz, EMG: 30–300 Hz) before being digitized at a sampling rate of 128 Hz and stored on a computer.
Sleep Deprivation
After baseline recording, animals were sleep deprived for 6 hours during the last half of the light phase (13:00 –19:00). For this purpose, animals and the EEG/EMG recordings for signs of sleep were continuously observed, and various objects (pieces of paper, plastic tubes) were introduced into the cage, or gentle handling was used to arouse the mouse as soon as it adapted a sleeping posture or showed EEG signs of sleep.12 At dark onset (19:00), sleep deprivation was terminated and animals were left to sleep freely while being recorded for 24 h (from 19:00 to 19:00 on the next day).
Cage Change
To examine the ability of mice to maintain wakefulness in a novel environment, we recorded the EEG after cage change.13,14 A new set of animals were used for cage change studies. Mice were allowed to adapt to recording and baseline sleep data were collected. Without change of LD cycle, mice were then transferred from their habitual home cage to clean cages with fresh bedding and new nesting material at 13:00, and sleep-wake behaviors were assessed over the next 12 hours after the cage change (from 13:00 to 01:00 on the next day) with food and water freely available.
Analysis of Sleep Data
After sleep data were collected, EEG/EMG records were scored semiautomatically by using the SleepSign scoring system (Kissei Comtec America, Irvine, CA) in 4-sec epochs scored as wake, REM sleep, and NREM sleep on the basis of standard criteria of rodent sleep.15–17 This preliminary scoring was visually inspected and corrected when appropriate. The distribution and amount of the behavioral states were analyzed by expressing them as a percentage of recording time. Awake or sleep bouts were defined as a period of each state that was initiated by 2 consecutive 4-sec epochs and terminated by 2 consecutive 4-sec epochs classified as a different state from that of the bout. Mean duration of each state during the light or dark period was calculated by dividing the total time by the number of corresponding bouts. EEG spectral power was tallied in 0.5 Hz bins using Fast Fourier Transformation (FFT) of each 4-sec epoch. To account for individual differences in the EEG signal, power density value for each frequency bin was expressed as a proportion of the mean of total power across all frequency bins for NREM or REM sleep (e.g., the mean power of 0.5–25 Hz over a 24 hour period).18 Delta power was determined by averaging the power density values from 1.0 to 4.0 Hz, and theta power was determined by averaging the power density values from 5.0 to 9.0 Hz. NREM and REM sleep latencies were calculated as the time from an intervention (cage change) to the first epoch of NREM or REM sleep. In addition, in order to determine differences from baseline sleep and sleep loss during sleep deprivation, we examined the percentage change relative to baseline value (100 × [treatment − baseline] / baseline) and the percentage sleep regained relative to the amount lost (100 × [recovery − baseline] / amount of sleep lost during sleep deprivation) for the recovery 12-h dark and recovery 12-h light periods after sleep deprivation (Figure 4D and E) and the 6-hr light period after cage change (Figure 5E).4,14
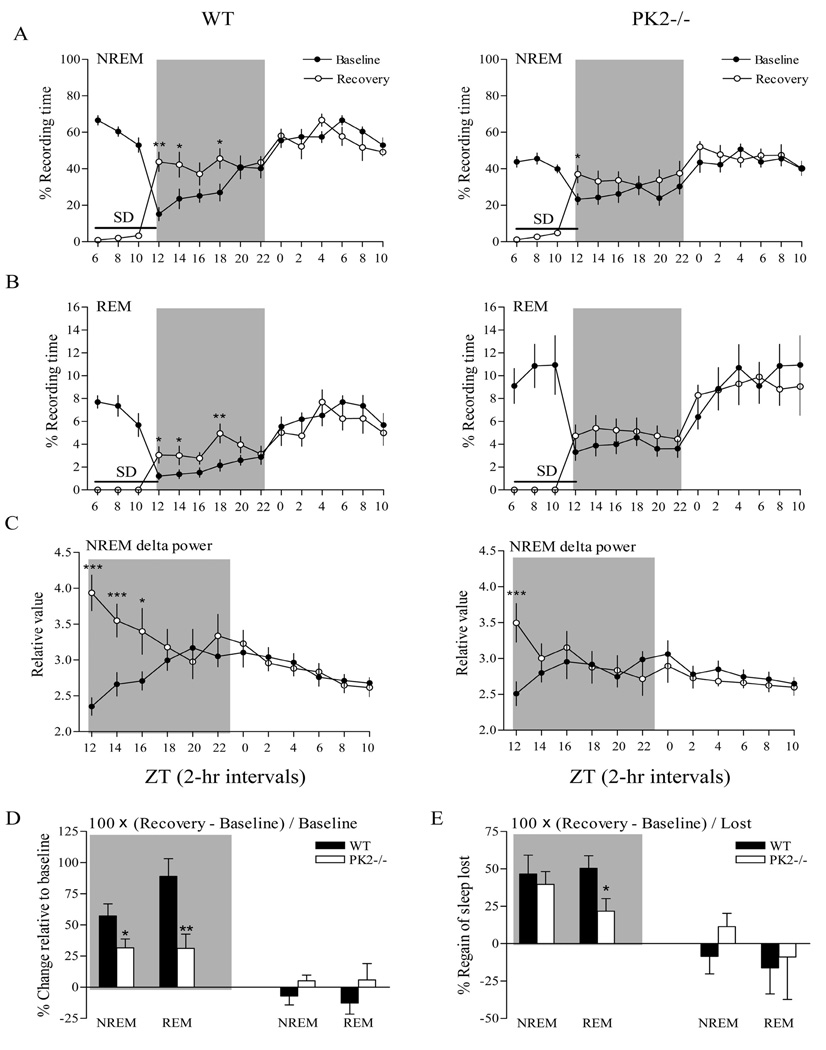
Figure 4.
Recovery from sleep deprivation in wild-type (WT) and PK2−/− mice. Animals were sleep deprived (SD) in the last 6 h of the light phase, as indicated with the horizontal line. The recovery phase began at dark onset (zeitgeber time, ZT 12) and continued throughout the 12-h dark period (indicated by the shaded background) and the 12-h light period. Left panels: Baseline (filled circles) and recovery (open circles) patterns in WT mice for NREM sleep (A), REM sleep (B), and NREM delta power (C), graphed in 2-hr intervals. Right panels (A, B, C): Baseline (filled circles) and recovery (open circles) patterns in PK2−/− mice. *P < 0.05; **P < 0.01; ***P < 0.001, post hoc Bonferroni test. D, Percentage change relative to baseline value (100 × [recovery − baseline] / baseline) for NREM and REM sleep during the 12-h dark period (indicated by the shaded background) and the 12-h light period after 6-h SD in WT and PK2−/− mice. *P < 0.05; **P < 0.01, unpaired t-test. E, Percentage sleep regained during recovery period relative to sleep lost during 6-h SD (100 × [recovery − baseline] / amount of sleep lost during SD) for NREM and REM sleep during the 12-h dark period (indicated by the shaded background) and the 12-h light period after 6-h SD in WT and PK2−/− mice. *P < 0.05; unpaired t-test. n = 9 mice / per genotype.
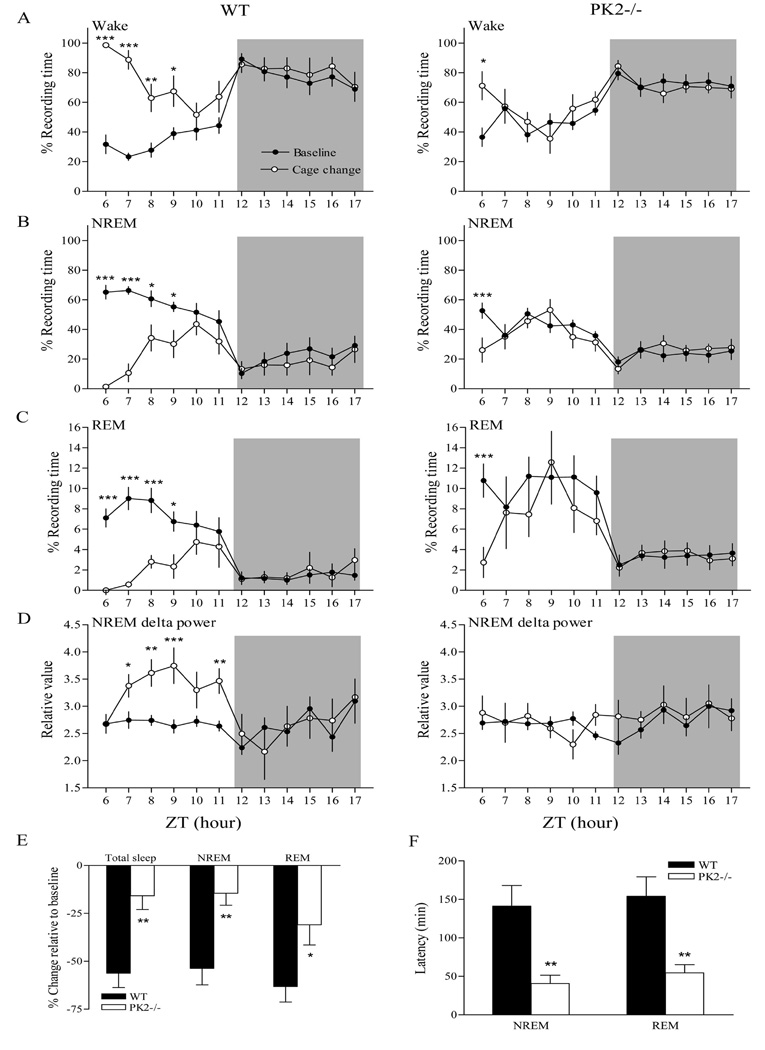
Figure 5.
Sleep-wake patterns after cage change in wild-type (WT) and PK2−/− mice. Animals were placed in new and clean cages at ZT6. Recordings were made from the last 6 h of the light phase to the first 6 h of the dark phase (indicated by the shaded background). Distribution of wake (A), NREM sleep (B), REM sleep (C) after cage change in 1-hr intervals for WT (Left panels) and PK2−/− mice (Right panels). *P < 0.05; **P < 0.01; ***P < 0.001, post hoc Bonferroni test. D, NREM delta power plotted hourly after cage change in WT (Left panels) and PK2−/− mice (Right panels). *P < 0.05; **P < 0.01; ***P < 0.001, post hoc Bonferroni test. E, Percentage change relative to baseline value (100 × [treatment − baseline] / baseline) for total sleep, NREM, and REM sleep during the 6-h light period after cage change in WT and PK2−/− mice. *P < 0.05; **P < 0.01, compared with WT mice, unpaired t-test. F, Latency of NREM sleep and REM sleep in minutes following cage change in WT and PK2−/− mice. **P < 0.01, compared with WT mice, unpaired t-test. n = 7 mice / per genotype.
Statistical Analysis
All results were expressed as means ± s.e.m. All main effects of the factors “genotype” (PK2−/− vs WT), “time” (baseline vs recovery, or baseline vs cage change) were analyzed by 2-way ANOVA for repeated measures. When main effects were present, post hoc Bonferroni tests were performed to evaluate further differences between genotypes. In a few instances, comparisons between WT and PK2−/− mice were determined using unpaired, two-tailed Student’s t tests.
RESULTS
Baseline Sleep-Wake Patterns under LD Cycles
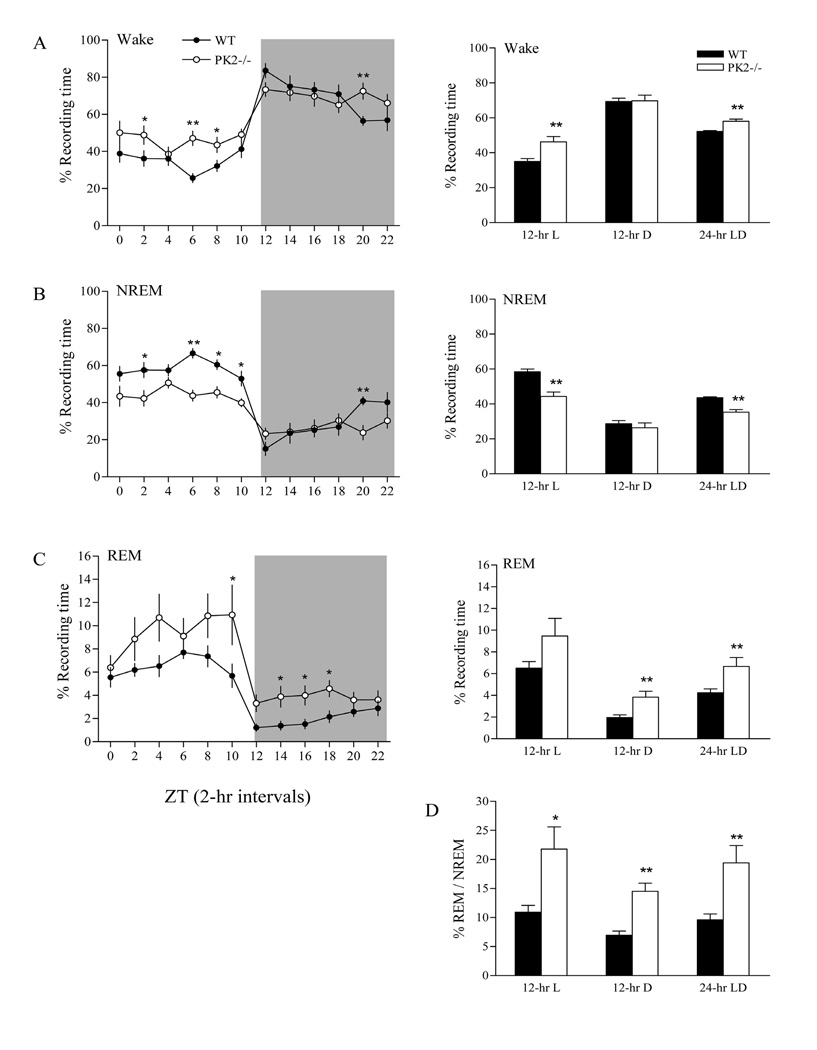
Sleep-wake patterns of PK2−/− mice differed from those of WT mice over the 24-hr baseline LD condition. PK2−/− mice slept about 83.5 min less than WT mice per day (genotype effect, F = 10.73, P < 0.01). The reduced total sleep time in PK2−/− mice resulted from changes in the 12-h light phase (− 80.5 min; genotype effect, F = 21.85, P < 0.0001), but not in the 12-h dark phase (− 3.0 min; genotype effect, F = 0.03, P = 0.87; Figure 1A). This altered sleep distribution resulted in a smaller light : dark ratio of sleep quantity in PK2−/− mice (Figure 1, left panels: A, B). The reduction in the total amount of sleep was entirely attributed to a reduction of the NREM sleep. PK2−/− mice had reduced NREM sleep time compared to WT mice in the 12-h light phase (− 101.9 min; genotype effect, F = 48.97, P < 0.0001), while in the 12 h-dark phase, PK2−/− mice had NREM sleep time similar to WT mice (− 16.5 min; genotype effect, F = 0.96, P = 0.33; Figure 1B). In contrast, PK2−/− mice had increased REM sleep time in both the 12-h light phase (+ 21.4 min; genotype effect, F = 12.62, P < 0.0001) and the 12-h dark phase (+ 13.5 min; genotype effect, F = 26.24, P < 0.0001; Figure 1C). Thus, deficiency of the PK2 gene affected NREM and REM sleep quantities in opposite directions. The REM / NREM sleep ratio was significantly higher in PK2−/− mice, during the 12-h light period, 12-h dark period, or the entire 24-h LD period (unpaired t-test, P < 0.05, 0.01, and 0.01, respectively; Figure 1D).
Figure 1.
Baseline sleep-wake patterns in wild-type (WT) and PK2−/− mice. Left panels: Distribution of wake (A), non-rapid eye movement (NREM) sleep (B) and rapid eye movement (REM) sleep (C) in 2-hr intervals under a 12 h : 12 h light : dark (LD) cycle for WT (filled circles) and PK2−/− (open circles) mice. Sleep-wake values were expressed as percentage of recording time. Recordings were initiated at light onset (zeitgeber time, ZT0) and the dark phase is indicated by the shaded background. *P < 0.05; **P < 0.01, post hoc Bonferroni test. Right panels: Amounts of baseline sleep-wake states (A: wake; B: NREM; C: REM) and ratio (%) of REM / NREM (D) were averaged over the 12-h light, 12 h-dark and 24-hr intervals for each genotype. *P < 0.05; **P < 0.01, unpaired t-test. Values represent group means ± s.e.m. n = 9 mice / per genotype
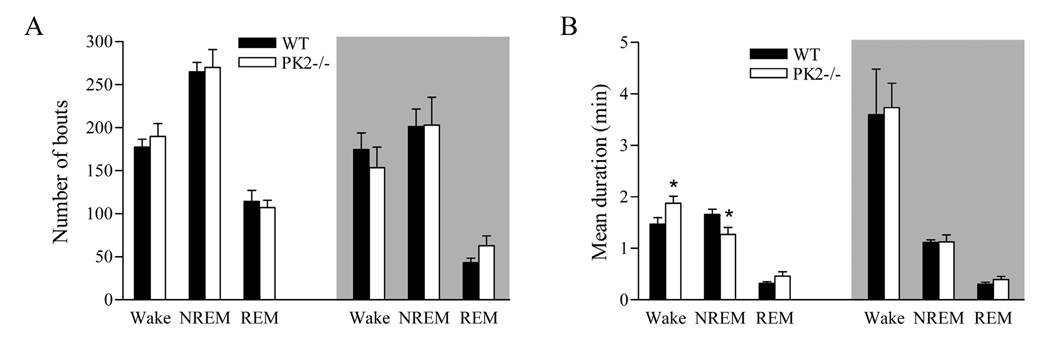
We further analyzed the sleep architecture of PK2−/− mice. The reduction of NREM sleep in PK2−/− mice in the 12-h light phase was mainly due to shorter mean duration of NREM sleep bouts (PK2−/−, 1.27 ± 0.13 min; WT, 1.66 ± 0.10 min; unpaired t-test, P < 0.05), without any change to the number of NREM sleep bouts (PK2−/−, 270.2 ± 20.7; WT, 264.8 ± 11.0). The mean duration (PK2−/−, 1.13 ± 0.13 min; WT, 1.11 ± 0.05 min) and the number (PK2−/−, 203.1 ± 32.4; WT, 201.3 ± 20.4) of NREM sleep bouts did not differ between genotypes in the 12-h dark phase, and there was no difference in the amount of NREM sleep in the dark phase (Figure 2). The increased wakefulness in PK2−/− mice in the 12-h light phase was due to a longer mean duration of bouts (PK2−/−,1.87 ± 0.13 min; WT, 1.47 ± 0.10 min; unpaired t-test, P < 0.05), whereas the number of wake bouts was not significantly different (PK2−/−, 189.9 ± 14.9; WT, 177.6 ± 9.2). The increased REM sleep in PK2−/− mice in the 12-h light phase may be due to increase of mean duration of REM sleep bouts (PK2−/−, 0.47 ± 0.08 min; WT, 0.31 ± 0.03 min; unpaired t-test, P = 0.08). The number of REM sleep bouts during light phase was similar between genotypes (PK2−/−, 107.3 ± 8.4; WT, 114.4 ± 12.8; unpaired t-test, P = 0.64). In the 12-h dark phase, PK2−/− mice showed trends of increase in both the number (PK2−/−, 62.9 ± 10.1; WT, 43.1 ± 5.0; unpaired t-test, P = 0.09) and the mean duration (PK2−/−, 0.39 ± 0.05 min; WT, 0.30 ± 0.03 min; unpaired t-test, P = 0.15) of REM sleep bouts.
Figure 2.
Baseline sleep-wake fragmentation in wild-type (WT) and PK2−/− mice. The number (A) and the mean duration (B) of wake, NREM sleep and REM sleep bouts during the light and dark (indicated by the shaded background) phases. *P < 0.05, unpaired t-test. n = 9 mice / per genotype.
EEG spectral power in the delta frequency range (1 – 4 Hz) is often used as an indicator for sleep drive or intensity.19 We thus quantified the EEG power in the PK2−/− and WT mice. Although PK2−/− mice had a decrease in the amount of total sleep, the distribution of NREM sleep delta power was not different between PK2−/− and WT mice in both 12-h light (genotype effect, F = 1.15, P = 0.29) and 12-h dark (genotype effect, F = 0.001, P = 0.97) phases (Figure 4C). REM sleep theta power is decreased in PK2−/− mice in both 12-h light phase (genotype effect, F = 33.05, P < 0.0001) and 12-h dark phase (genotype effect, F = 22.35, P < 0.0001), which is consistent with increased REM sleep time in PK2−/− mice.
Sleep in Constant Dark Conditions
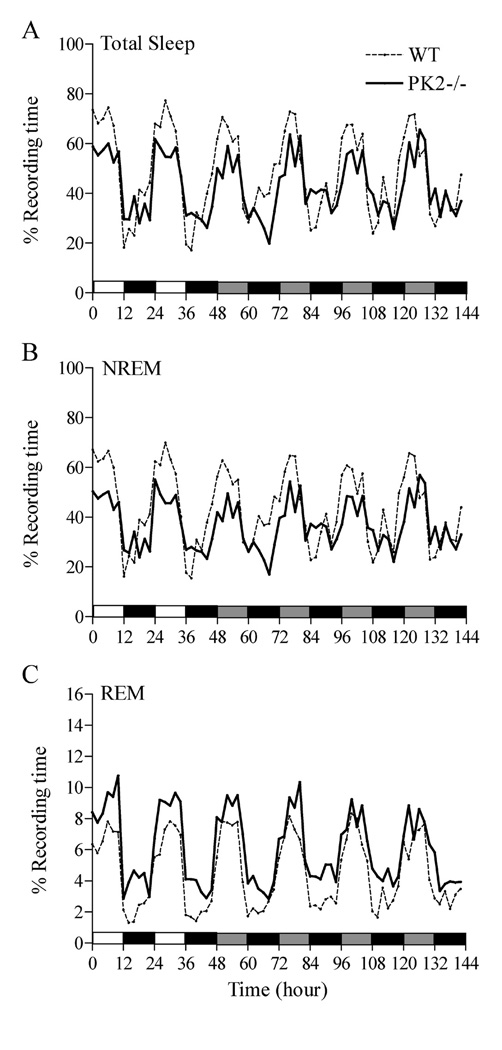
Following 2 days under LD conditions, animals were continually recorded for 4 more days under constant dark (DD) conditions. As shown in Table 1, PK2−/− mice spent less time in total sleep and NREM sleep than WT mice during the light phase under LD and the subjective light phase under DD. As under the LD condition, PK2−/− mice spent more time in REM sleep during the dark phase under DD (Table 1). Distributions of total sleep, NREM, and REM sleep under the 2 days of LD and 4 days of DD were shown in Figure 3. ANOVA tests revealed there were significant genotype differences between PK2−/− and WT mice under DD condition in total sleep (genotype effect, F = 10.48, P < 0.001), NREM sleep (genotype effect, F = 36.43, P < 0.001), and REM sleep (genotype effect, F = 45.27, P < 0.001).
Table 1.
Sleep Amounts for 12-hr Interval in LD and DD Conditions
| Total Sleep |
NREM |
REM |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | PK2−/− | P | WT | PK2−/− | P | WT | PK2−/− | P | |
| LD1 | |||||||||
| Light | 70.7 ± 1.8 | 56.8 ± 3.7 | <0.01 | 64.4 ± 2.0 | 47.9 ± 3.2 | <0.01 | 6.3 ± 0.3 | 8.9 ± 1.4 | 0.09 |
| Dark | 33.5 ± 2.4 | 36.5 ± 2.4 | 0.37 | 31.0 ± 2.3 | 31.3 ± 1.9 | 0.81 | 2.4 ± 0.3 | 5.2 ± 1.0 | <0.05 |
| LD2 | |||||||||
| Light | 65.4 ± 2.4 | 52.9 ± 3.4 | <0.01 | 59.3 ± 2.3 | 45.2 ± 2.9 | <0.01 | 6.2 ± 0.4 | 7.8 ± 1.2 | 0.23 |
| Dark | 30.9 ± 1.6 | 32.7 ± 2.4 | 0.53 | 28.6 ± 1.4 | 28.1 ± 2.1 | 0.97 | 2.3 ± 0.3 | 4.6 ± 0.8 | <0.05 |
| DD1 | |||||||||
| Subjective Light | 61.9 ± 3.1 | 49.1 ± 4.9 | <0.05 | 55.4 ± 2.9 | 41.2 ± 4.5 | <0.05 | 6.5 ± 0.7 | 7.9 ± 0.8 | 0.21 |
| Subjective Dark | 35.8 ± 4.3 | 29.8 ± 5.7 | 0.38 | 33.7 ± 4.2 | 25.6 ± 5.0 | 0.23 | 2.0 ± 0.5 | 4.1 ± 0.8 | <0.05 |
| DD2 | |||||||||
| Subjective Light | 61.2 ± 2.3 | 50.7 ± 3.9 | <0.05 | 54.9 ± 2.8 | 43.1 ± 4.3 | <0.05 | 6.3 ± 0.9 | 7.7 ± 0.8 | 0.26 |
| Subjective Dark | 34.4 ± 5.1 | 38.8 ± 3.2 | 0.46 | 31.8 ± 4.9 | 34.1 ± 2.8 | 0.63 | 2.5 ± 0.6 | 4.7 ± 0.7 | <0.05 |
| DD3 | |||||||||
| Subjective Light | 60.9 ± 3.5 | 49.6 ± 3.2 | <0.05 | 54.8 ± 2.8 | 42.3 ± 2.8 | <0.01 | 6.2 ± 0.5 | 7.3 ± 1.3 | 0.46 |
| Subjective Dark | 32.9 ± 4.7 | 35.3 ± 5.2 | 0.74 | 30.4 ± 4.2 | 30.6 ± 4.2 | 0.92 | 2.5 ± 0.7 | 4.7 ± 1.2 | 0.14 |
| DD4 | |||||||||
| Subjective Light | 61.9 ± 2.6 | 53.1 ± 3.8 | 0.08 | 55.7 ± 2.4 | 45.9 ± 3.6 | <0.05 | 6.3 ± 0.6 | 7.2 ± 0.9 | 0.39 |
| Subjective Dark | 32.5 ± 4.6 | 35.7 ± 4.6 | 0.73 | 29.8 ± 4.1 | 31.2 ± 3.9 | 0.76 | 2.7 ± 0.8 | 4.5 ± 1.0 | 0.19 |
Data are presented as mean ± SEM of 12-hr intervals for 2 days under light / dark and 4 days of constant dark. P indicates the probability of the unpaired t tests comparing the genotypes. (n = 7 mice / per genotype)
Figure 3.
Time courses of total sleep (A), NREM sleep (B) and REM sleep (C) for 2 d baseline under 12 h : 12 h light : dark (LD) and followed by 4 d under constant darkness (DD) in wild-type (WT) and PK2−/− mice. Black horizontal bars indicate dark phase, and shaded bars indicate the subjective light phases under DD. n = 7 mice / per genotype.
Sleep Deprivation and Recovery
Animals were sleep deprived for 6 hours in the second half of the light phase. Although brief NREM sleep bouts could not be prevented, especially toward the end of SD, the total amount of residual NREM sleep during the SD period were similar in PK2−/− and WT mice. The recovery period began at the onset of the dark phase and persisted throughout the 12-h dark period and the subsequent 12-h light period (Figure 4). Compared with the same period under baseline LD conditions, NREM sleep quantity was significantly increased in the recovery 12-h dark period immediately after SD in both WT (condition effect, F = 30.94, P < 0.0001) and PK2−/− mice (condition effect, F = 18.81, P < 0.0001; Figure 4A). Similarly, REM sleep time was also significantly increased in the recovery period in WT (condition effect, F = 33.16, P < 0.0001) and PK2−/− mice (condition effect, F = 13.30, P < 0.001; Figure 4B). Post hoc tests revealed significant increases in both NREM and REM sleep amount during hours 1 to 4 and 7 to 8 of the 12-h recovery dark period in WT mice. However, an increased NREM sleep quantity was observed only during hours 1 and 2 of the 12-h recovery dark period in PK2−/− mice, and no significant increase in REM sleep time was found in any of the 2-h intervals. In the recovery 12-h light period, neither NREM nor REM sleep amount differed from the corresponding baseline levels in WT or PK2−/− mice (Figures 4A and B).
Direct comparison of recovery sleep between the two genotypes was complicated by different baseline levels and the different amounts of sleep lost during SD in PK2−/− and WT mice. For this reason, we analyzed the recovery sleep response by two different criteria for the recovery 12 h dark period as described.4 For criterion 1, we determined the percentage change in recovery sleep relative to the baseline level (100 × [recovery − baseline] / baseline]). As shown in Figure 4D, PK2−/− mice had a significantly smaller change in NREM sleep amount relative to the baseline value than WT mice in the recovery 12-h dark period (PK2−/−, 31.55% ± 7.16 %; WT, 57.23% ± 9.61 %; unpaired t-test, P < 0.05), as was for REM sleep (PK2−/−, 31.23% ± 11.48 %; WT, 88.92% ± 14.30 %; unpaired t-test, P < 0.01). For criterion 2, we calculated the percentage of regained sleep relative to the lost sleep amount during the 6-h period of SD (100 × [recovery − baseline] / amount of sleep lost during SD). REM sleep rebound in PK2−/− mice was reduced (21.74% ± 8.41 %) compared with WT mice (50.39% ± 8.42 %; unpaired t-test, P < 0.05), while there was no significant difference between genotypes in the amount of regained NREM sleep (PK2−/−, 39.61% ± 8.55 %; WT, 46.63% ± 12.57 %; unpaired t-test, P > 0.05) during the recovery 12-h dark period (Figure 4E). During the recovery 12-h light period after SD, however, no significant differences were found in NREM and REM sleep amount between WT and PK2−/− mice, according to both criteria (Figure 4D, E). Taken together, these results revealed that the sleep rebound response of PK2−/− mice to sleep deprivation was attenuated.
We next examined the delta power change in response to sleep deprivation. As shown in Figure 4C, NREM sleep delta power was significantly increased in both the PK2−/− and WT mice during the immediate recovery period after SD, indicating an increase in the drive for sleep. Interestingly, post hoc tests revealed that the increase in NREM delta power was restricted to the first 2 hours of the recovery period in PK2−/− mice, whereas in WT mice the increase lasted for 6 hours. Compared with the respective baseline values, the percentage of NREM sleep delta power increase in PK2−/− mice was also significantly smaller than that of WT mice (genotype effect, F = 9.85, P < 0.01) during the recovery 12-h dark period after SD. These results indicate that PK2−/− mice displayed attenuated rebound in NREM sleep delta power to sleep deprivation.
Sleep after Cage Change
Over the course of animal handling, we noticed a faster return of the PK2−/− mice to rest during the light period in response to cage changes. We thus quantified this observation by direct sleep measurement. When placed into new cages with clean bedding during the light phase, WT and PK2−/− mice showed increases in wakefulness and decreases in NREM and REM sleep amounts (Figure 5A, B, and C). During the 6-h period after cage change, considerable changes in wakefulness (condition effect, F = 57.71, P < 0.0001), NREM sleep (condition effect, F = 55.97, P < 0.0001), and REM sleep amount (condition effect, F = 58.52, P < 0.0001) were observed in WT mice, while only NREM (condition effect, F = 5.18, P < 0.05) and REM sleep (condition effect, F = 14.23, P < 0.01) quantities changed significantly in PK2−/− mice. Wakefulness increase by cage change was only evident in the first hour in PK2−/− mice, however, ANOVA analysis indicated no significant differences within the total 6-h period after cage change (condition effect, F = 1.63, P = 0.21). Compared to their respective baseline levels, WT mice showed a significant increase in wakefulness and decreases in NREM and REM sleep amount from 1 to 4 hours after cage change; however, PK2−/− mice only showed such changes for the first hour after cage change.
Comparing the percentage change in sleep measures relative to baseline levels during the first 6-h period after cage change (100 × [treatment − baseline] / baseline), PK2−/− mice showed significantly smaller reductions in total sleep (PK2−/−, 15.8% ± 7.2%; WT, 56.2% ± 7.5%; unpaired t-test, P < 0.01), NREM sleep (PK2−/−, 14.5% ± 6.2%; WT, 53.7% ± 8.6%; unpaired t-test, P < 0.01) and REM sleep (PK2−/−, 30.9% ± 10.6%; WT, 63.7% ± 8.1%; unpaired t-test, P < 0.05; Figure 5E). Consistent with these data, NREM sleep delta power significantly increased in WT mice during the light period after cage change (condition effect, F = 53.34, P < 0.0001); however, no such difference was observed in PK2−/− mice (condition effect, F = 1.14, P = 0.29; Figure 5D). In addition, we also analyzed the time from cage change to the first epoch of sleep (i.e., sleep latency). As shown in Figure 5F, PK2−/− mice had shorter NREM and REM sleep latencies after cage change than did WT mice. These results indicated that PK2−/− mice had impaired abilities of maintaining wakefulness in response to novel environments.
DISCUSSION
PK2 Deficiency Affects Amount of Sleep under Both LD and DD Conditions
The most striking finding of this work is that PK2−/− mice slept less per day than WT mice under a LD cycle. The reduced sleep time in PK2−/− mice occurred mainly during the light period and was entirely attributed to a reduction of NREM sleep time and a shorter duration of NREM sleep bouts. Interestingly, REM sleep was increased in PK2−/− mice during both light and dark periods, even though the amount of total sleep was decreased. The alterations in total sleep, NREM and REM sleep in PK2−/− mice were preserved under DD condition, indicating that these alterations were not caused by masking effects of the LD cycle. Thus, the mutation of the PK2 gene influences both the distribution and the amount of sleep, implying that PK2 is a molecule that regulates both circadian and homeostatic processes of sleep.
It has been proposed that the SCN is a component of the wake-regulatory pathway and that the efferent signals from the SCN have arousal-promoting effects in diurnal animals.20 The reduced NREM sleep in PK2−/− mice occurred primarily in the light phase but not in the dark phase, and this phase-dependent effect is consistent with rhythmic expression of PK2 mRNA in the SCN.7 This result implies the involvement of PK2 in sleep-wake regulation is dependent on its circadian effect of PK2. A correlation between increased daytime locomotor activity and reduction of the molecular rhythm of PK2 in the SCN has been observed in transgenic mice expressing a mutant Huntington’s gene.21 The SCN is connected to hypothalamic and brainstem nuclei having important roles in sleep-wake regulation.22 The dorsomedial hypothalamus, a major SCN target nucleus, may integrate circadian signals from the SCN and other neural inputs while projecting to other sleep-wake regulatory regions, such as the dorsal tuberomammillary nucleus, the locus coeruleus, the ventrolateral preoptic nucleus and the median preoptic nucleus.23–25 PKR2, which encodes the receptor for PK2, is expressed at high levels in the dorsomedial hypothalamus,10 consistent with a possible involvement of PK2 in sleep-wake regulation via this relay nucleus.
REM sleep was increased in PK2−/− mice during both light and dark periods, and PK2−/− mice had different homeostatic control of sleep during the recovery dark period after sleep deprivation, suggesting that at least some function of PK2 in sleep regulation might be independent of SCN circadian clock function. Besides the SCN, PK2 is also expressed in other brain regions,10 such as in the medial preoptic nucleus, the amygdala and nucleus accumbens that have been implicated in sleep control,26–28 and therefore may contribute to sleep changes in PK2−/− mice. In addition, PKR2 is also expressed in the nuclei that are not SCN target areas but that are important for sleep-wake regulation,10 such as the lateral hypothalamus, the perifornical region of the hypothalamus and the dorsal raphe nucleus.29–31 It is possible that the changes of sleep regulation in PK2−/− mice, such as those affecting REM sleep, may be due to the functional changes of these areas in the PK2−/− mice.
Mutation of the PK2 Gene Attenuated the Compensatory response to sleep deprivation
Loss of circadian genes affects the homeostasis of sleep, as displayed by their altered response to sleep deprivation. Deletion of Bmal1/Mop3 caused an attenuated compensatory response to sleep deprivation,4 where as Clock mutant mice presented a decreased rebound after sleep deprivation in REM sleep but not NREM sleep.3 Similar to Clock mutant mice, DBP knockout mice showed a decreased rebound in REM sleep after sleep deprivation, without any difference in NREM sleep rebound.6 Cry1 and Cry2 double deficient mice showed no rebound in the amount of NREM and REM sleep after sleep deprivation.5 Per1 and Per2, negative elements of the molecular clock in the SCN, however, may not be required for homeostatic regulation of the daily amount of wakefulness, NREM sleep, or REM sleep.12 In the present study, PK2−/− mice showed a reduced rebound response in REM sleep, NREM sleep, and NREM sleep delta power in response to sleep deprivation. PK2−/− mice generated compensatory increases in NREM and REM sleep times and NREM delta power during the recovery 12-h dark period, but the rebound sleep durations were shorter than in WT mice. Unlike WT mice, PK2−/− mice exhibited a smaller change relative to baseline values in NREM and REM sleep times and NREM delta power during recovery 12-h dark period. Moreover, during the recovery 12-h dark period, PK2−/− mice regained a smaller percentage of the total amount of REM sleep lost, although they regained a similar percentage of the lost NREM sleep. Thus, PK2−/− mice showed an attenuated compensatory response to sleep deprivation, further supporting the idea that PK2 has an influence on the homeostatic regulation of sleep.
EEG delta power is considered a prominent marker of NREM sleep homeostatic regulation, as it exhibits a predictive quantitative relationship with the duration of previous wakefulness.19 EEG delta power is increased in mice lacking Bmal1/Mop3,4 Cry1 and Cry2,5 whereas the daily amplitude of EEG delta power decreases in mice lacking the DBP gene.6 No difference in NREM sleep delta power was found between PK2−/− and WT mice during both baseline light and dark periods, even though the PK2−/− mice slept less daily than did WT mice. A possible explanation is that mutation of the PK2 gene slows the accumulation of homeostatic sleep parameters, e.g., sleep “need”. The same finding was reported in Clock mutant mice.3 Immediately after sleep deprivation, although NREM sleep delta power increased dramatically in both PK2−/− and WT animals, the PK2−/− mice had a much shorter rebound duration and a smaller increase of NREM sleep delta power in the recovery 12-h dark period, indicating that NREM sleep delta power rebound is significantly attenuated in PK2−/− mice. For NREM sleep delta power, a decreased compensatory response following sleep deprivation was also seen in mice lacking Cry1 and Cry2, Per1 and Per2, and Bmal1/Mop3.4,5,32 The altered response to sleep deprivation in these mutant mice of circadian genes appears to support the notion that the circadian and homeostatic processes underlying the regulation of sleep are linked, at least at the molecular level.
PK2−/− Mice Had Impaired Abilities of Maintaining Wakefulness in Response to Novel Environments
Considerable evidence supports that environmental challenges can have significant effects on arousal states in animals, including prolonged sleep latencies and reduced total sleep time.13,14 When confronted with behavioral challenges such as novel housing condition and starvation, it is important for an animal to maintain an elevated alertness state, at least for a short period of time. Such a wakefulness maintenance response may be required for proper adaptation of animals to these environmental challenges. In the present experiment, we show that the PK2−/− mice did not exhibit prolonged wakefulness in response to cage change. During the first 6-h period after cage change, the PK2−/− mice showed a substantially reduced increase in wakefulness. The enhancement of wakefulness by cage change was restricted to only the first hour in PK2−/− mice, whereas it lasted for 4 hours in WT mice. These results indicated that a null mutation of the PK2 gene attenuates the arousal enhancement induced by cage change. This reduced response is unlikely to be due to a difference in the baseline sleep parameters, as PK2−/− mice actually had reduced total sleep amount during light phase. The neurobiological mechanism of PK2 in meditating the arousal stimulation remains to be determined. It is well established that amygdala is a critical structure for stress responsiveness and emotional regulation.33 The amygdala is heavily interconnected with hypothalamic and brainstem nuclei known to be involved in sleep control.34 Therefore, the attenuated response of PK2−/− mice to novel environments may be linked to the expression of PK2 in the amygdala.10
In summary, mice lacking the PK2 gene, which encodes a postulated SCN output molecule, exhibit altered baseline sleep parameters and an attenuated compensatory rebound in response to sleep deprivation, in addition to an altered circadian sleep distribution. PK2−/− mice also exhibit impaired response to behavioral challenges. These findings indicate PK2 is involved in both circadian and homeostatic regulation of sleep.
ACKNOWLEDGEMENTS
We thank Chongwei Wen for help with mouse genotyping and members of Zhou laboratory for critical comments on the manuscript. This work was supported in part by grants from National Institutes of Health and Kinexis Inc (to QYZ)
Disclosure Statement
The main funding for this work is from NIH. A grant from Kinexis Inc. supported the early phase of this work. Dr. Zhou is a co-founder of Kinexis Inc. and a member of its scientific advisory board. Drs. Hu, Li, Zhang, Boehmer, and Siegel have indicated no conflicts of interest.
REFERENCES
- 1.Borbely AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 2.Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic nucleus : the mind's clock. Oxford University Press; 1991. [Google Scholar]
- 3.Naylor E, Bergmann BM, Krauski K, et al. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 5.Wisor JP, O’Hara BF, Terao A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20–34. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franken P, Lopez-Molina L, Marcacci L, Schibler U, Tafti M. The transcription factor DBP affects circadian sleep consolidation and rhythmic EEG activity. J Neurosci. 2000;20:617–625. doi: 10.1523/JNEUROSCI.20-02-00617.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng MY, Bullock CM, Li C, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 8.Zhou QY, Cheng MY. Prokineticin 2 and circadian clock output. Febs J. 2005;272:5703–5709. doi: 10.1111/j.1742-4658.2005.04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng MY, Bittman EL, Hattar S, Zhou QY. Regulation of prokineticin 2 expression by light and the circadian clock. BMC Neurosci. 2005;6:17–27. doi: 10.1186/1471-2202-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng MY, Leslie FM, Zhou QY. Expression of prokineticins and their receptors in the adult mouse brain. J Comp Neurol. 2006;498:796–809. doi: 10.1002/cne.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JD, Hu WP, Boehmer L, et al. Attenuated circadian rhythms in mice lacking the prokineticin 2 gene. J Neurosci. 2006;26:11615–11623. doi: 10.1523/JNEUROSCI.3679-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiromani PJ, Xu M, Winston EM, Shiromani SN, Gerashchenko D, Weaver DR. Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes. Am J Physiol Regul Integr Comp Physiol. 2004;287:R47–R57. doi: 10.1152/ajpregu.00138.2004. [DOI] [PubMed] [Google Scholar]
- 13.Mochizuki T, Crocker A, McCormack S, Yanagisawa M, Sakurai T, Scammell TE. Behavioral state instability in orexin knock-out mice. J Neurosci. 2004;24:6291–6300. doi: 10.1523/JNEUROSCI.0586-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Xiao J, Parris BS, Fang J, Sanford LD. Differential effects of two types of environmental novelty on activity and sleep in BALB/cJ and C57BL/6J mice. Physiol Behav. 2005;85:419–429. doi: 10.1016/j.physbeh.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Kim D, Shin HS. Lack of delta waves and sleep disturbances during non-rapid eye movement sleep in mice lacking alpha1G-subunit of T-type calcium channels. Proc Natl Acad Sci U S A. 2004;101:18195–18199. doi: 10.1073/pnas.0408089101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinomiya K, Shigemoto Y, Omichi J, Utsu Y, Mio M, Kamei C. Effects of three hypnotics on the sleep-wakefulness cycle in sleep-disturbed rats. Psychopharmacology (Berl) 2004;173:203–209. doi: 10.1007/s00213-003-1727-0. [DOI] [PubMed] [Google Scholar]
- 17.Morrow JD, Opp MR. Sleep-wake behavior and responses of interleukin-6-deficient mice to sleep deprivation. Brain Behav Immun. 2005;19:28–39. doi: 10.1016/j.bbi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Parmentier R, Ohtsu H, Djebbara-Hannas Z, Valatx JL, Watanabe T, Lin JS. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci. 2002;22:7695–7711. doi: 10.1523/JNEUROSCI.22-17-07695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daan S, Beersma DG, Borbely AA. Timing of human sleep: recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246:R161–R183. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- 20.Edgar DM, Dement WC, Fuller CA. Effect of SCN lesions on sleep in squirrel monkeys: evidence for opponent processes in sleep-wake regulation. J Neurosci. 1993;13:1065–1079. doi: 10.1523/JNEUROSCI.13-03-01065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. Disintegration of the sleep-wake cycle and circadian timing in Huntington’s disease. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace-Schott EF, Hobson JA. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nat Rev Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 23.Aston-Jones G, Chen S, Zhu Y, Oshinsky ML. A neural circuit for circadian regulation of arousal. Nat Neurosci. 2001;4:732–738. doi: 10.1038/89522. [DOI] [PubMed] [Google Scholar]
- 24.Sutcliffe JG, de Lecea L. The hypocretins: setting the arousal threshold. Nat Rev Neurosci. 2002;3:339–349. doi: 10.1038/nrn808. [DOI] [PubMed] [Google Scholar]
- 25.Deurveilher S, Semba K. Indirect projections from the suprachiasmatic nucleus to major arousal-promoting cell groups in rat: implications for the circadian control of behavioural state. Neuroscience. 2005;130:165–183. doi: 10.1016/j.neuroscience.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 26.John J, Kumar VM. Effect of NMDA lesion of the medial preoptic neurons on sleep and other functions. Sleep. 1998;21:587–598. doi: 10.1093/sleep/21.6.587. [DOI] [PubMed] [Google Scholar]
- 27.Callaway CW, Henriksen SJ. Neuronal firing in the nucleus accumbens is associated with the level of cortical arousal. Neuroscience. 1992;51:547–553. doi: 10.1016/0306-4522(92)90294-c. [DOI] [PubMed] [Google Scholar]
- 28.Jha SK, Ross RJ, Morrison AR. Sleep-related neurons in the central nucleus of the amygdala of rats and their modulation by the dorsal raphe nucleus. Physiol Behav. 2005;86:415–426. doi: 10.1016/j.physbeh.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Abrams JK, Johnson PL, Hollis JH, Lowry CA. Anatomic and functional topography of the dorsal raphe nucleus. Ann NY Acad Sci. 2004;1018:46–57. doi: 10.1196/annals.1296.005. [DOI] [PubMed] [Google Scholar]
- 30.Gerashchenko D, Shiromani PJ. Different neuronal phenotypes in the lateral hypothalamus and their role in sleep and wakefulness. Mol Neurobiol. 2004;29:41–59. doi: 10.1385/MN:29:1:41. [DOI] [PubMed] [Google Scholar]
- 31.Willie JT, Chemelli RM, Sinton CM, Yanagisawa M. To eat or to sleep? Orexin in the regulation of feeding and wakefulness. Annu Rev Neurosci. 2001;24:429–458. doi: 10.1146/annurev.neuro.24.1.429. [DOI] [PubMed] [Google Scholar]
- 32.Kopp C, Albrecht U, Zheng B, Tobler I. Homeostatic sleep regulation is preserved in mPer1 and mPer2 mutant mice. Eur J Neurosci. 2002;16:1099–1106. doi: 10.1046/j.1460-9568.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 33.Nikolaev E, Kaczmarek L, Zhu SW, Winblad B, Mohammed AH. Environmental manipulation differentially alters c-Fos expression in amygdaloid nuclei following aversive conditioning. Brain Res. 2002;957:91–98. doi: 10.1016/s0006-8993(02)03606-5. [DOI] [PubMed] [Google Scholar]
- 34.Morrison AR, Sanford LD, Ross RJ. The amygdala: a critical modulator of sensory influence on sleep. Biol Signals Recept. 2000;9:283–296. doi: 10.1159/000014652. [DOI] [PubMed] [Google Scholar]