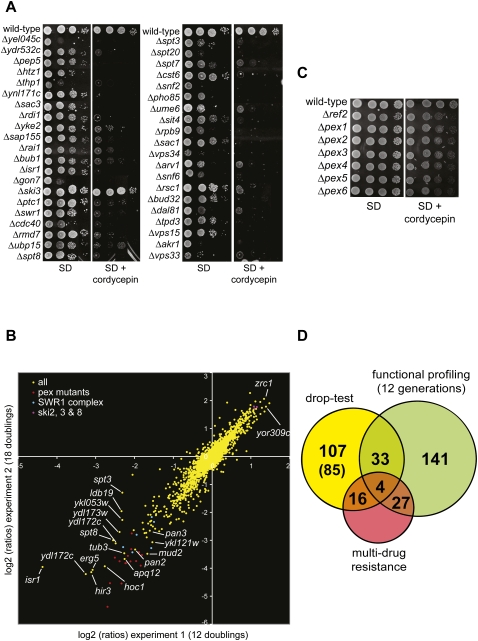
FIGURE 5.
Identification of cordycepin-responsive haploid deletion mutants. (A) The growth phenotypes of strains identified by drop-test screening were analyzed on medium containing or lacking 40 μg/mL cordycepin as indicated. Strains were grown for 3 d at 30°C and photographed. (B) Pools of all viable haploid deletion mutants from the systematic deletion library were grown in complete liquid medium with or without 20 μg/mL cordycepin for 12 or 18 generations (Experiments 1 and 2, respectively) by serial dilutions. Cells were then collected, the genomic DNA was extracted, and the barcode sequence tags associated with each gene deletion were amplified by PCR and hybridized on barcode DNA arrays as described (Decourty et al. 2008). The figure plots for each experiment (Experiment 1 on the x-axis and Experiment 2 on the y-axis) the log2 of the ratio for each deletion mutant of the barcode signal between the cultures without and with cordycepin (computed as in Decourty et al. 2008). Negative log2 (ratios) indicate strains more sensitive to cordycepin than the average population of mutants. Mutants in PEX genes, SWR1 complex genes, and SKI 2, 3, and 8 genes are color-coded as indicated. Arrows and labels point to mutants with strong phenotypes. (C) Cordycepin-sensitive growth phenotypes of pex mutants. The indicated strains were analyzed as described in A. (D) Graph depicting the overlap of mutants identified in drop-test screening versus functional profiling (after 12 generations). Also indicated are the numbers of multi-drug-resistance genes that were identified with the individual procedures or which are shared, respectively. In brackets the number of strains are indicated that are identified in the drop-test screening that could not be analyzed by functional profiling due to slow growth of the mutants.