Abstract
The chain length of Streptococcus pneumoniae type 3 capsular polysaccharide (cellubiuronic acid) is tightly regulated by the cellubiuronic acid synthase through an assembly process involving a catalytic motif that is potentially conserved over a wide range of related processive β-glucan synthases. Cellubiuronic acid is initiated on a lipid and is composed of alternating β-1,3-Glc and β-1,4-glucuronic acid (GlcUA) linkages. The entire assembly process is carried out by a polypeptide synthase thought to contain a single active site, suggesting that the donor specificity is controlled by the terminal nonreducing sugar in the acceptor subsite. Shortly after initiation, the synthase undergoes an allosteric transition accompanied by the tight binding of the nascent chain via its nonreducing oligosaccharide terminal segment to the carbohydrate acceptor recognition site. The chain length of polysaccharide assembled by recombinant synthase in Escherichia coli membranes was determined by an ejection mechanism that appeared to be a reversal of the allosteric transition of the synthase from the transitory to the fully processive state. The rates of both ejection and transition were shown to be highly sensitive to the concentration of UDP-GlcUA. As the concentration of UDP-GlcUA was increased, both the rate of synthesis and the processive turnover time increased. The product of the processive turnover time and the rate of synthesis predicted a marked increase in polysaccharide chain size (from 50 to 1150 kDa) over a relatively narrow concentration range of 1–11.5 μm UDP-GlcUA. The kinetic model chain length predictions were in close agreement with chemically determined sizes of polysaccharides synthesized at the same UDP-sugar concentrations. The model indicates that translocation occurs following the addition of GlcUA to the chain terminus, whereas UDP-Glc drives chain termination when inadequate levels of UDP-GlcUA are present. In sum, type 3 synthase appears to modulate polysaccharide chain length by functioning as a concentration-dependent kinetic timing device.
Control of the chain length of capsular polysaccharide is important in enabling streptococcal species to colonize and survive in a diversity of host physiological environments (1–3). The regulation of polysaccharide chain length in the biosynthesis of a wide variety of carbohydrate biopolymers has long been of interest. An early suggestion was that specific sugar modifications might serve as a chain termination signal in proteoglycan biosynthesis (4) or that chain length was determined by the ratio of synthase activity to primer concentration (5). Recently, chain termination and modal distribution of the lipopolysaccharide O8 and O9/O9a polymannans in Escherichia coli have been shown to be regulated by nonreducing end methylation (6), and O-antigen chain length in Shigella flexneri has been reported to be regulated by transcriptional control of the polymerase-encoding gene, wzy (7). Of particular relevance to the Streptococcus pneumoniae cellubiuronan, which grows by nonreducing end terminal glycosylation (8, 35), is the suggestion that chain length in heparin biosynthesis might be modulated by competition of the two nucleotide sugar substrates, UDP-N-acetylglucosamine and UDP-GlcUA,2 at the glycosyltransferase binding sites (9).
The cytosolic catalytic domain of cellubiuronan synthase contains the signature DDDQXRRW motif also found in the polymerases that assemble cellulose, chitin, hyaluronan, and other related β-glycans (10, 11), suggesting a similar inverting transfer mechanism for these members of the G2 glycosyltransferase family (12). Cellubiuronan formation proceeds by the alternate additions of β-1,3Glc and β-1,4GlcUA in an initial transitory processive stage followed by a fully processive stage. The fully processive polymerization reaction involves a tight association of the growing polysaccharide chain with the enzyme-carbohydrate recognition site of the type 3 synthase, except for a brief period during the translocation step of each catalytic cycle (13). Chain termination is modulated by a UDP-sugar-activated ejection process that appears to be an abortive translocation of the polysaccharide that occurs when insufficient levels of the UDP sugar precursors are available to maintain the polymerization reaction (14). In previous studies with S. pneumoniae membranes, UDP-Glc activated the ejection of preformed polysaccharide from the synthase with an apparent Km of 880 μm, and UDP-GlcUA inhibited this ejection with a Ki of 2 μm (14). Further, UDP-Glc-activated ejection of preformed polysaccharide was almost completely prevented by increasing the concentration of UDP-GlcUA to 10 μm. These data suggested that varying the concentration of UDP-GlcUA between ∼2 and 20 μm during the normal biosynthetic process, with UDP-Glc maintained at a high concentration, would allow the cells to regulate chain length over a large range in size.
A recent investigation in this laboratory revealed that UDP-Glc is present in the S. pneumoniae cytoplasm at much higher concentrations than UDP-GlcUA, which is present at barely detectable levels (15). In this study, we examined polysaccharide chain length modulation by recombinant synthase in E. coli membranes incubated with 1 mm UDP-Glc and low concentrations of UDP-GlcUA. In contrast with S. pneumoniae membranes, cellubiuronan synthase in E. coli membranes is not associated with preformed polysaccharide, allowing for uniform chain synthesis on a lipid primer (16, 17). Ejection of the polysaccharide chain, as described above, was previously studied in isolation from the assembly process. In the current investigation, polysaccharide assembly has been evaluated under steady state reaction conditions, wherein chain initiation, extension, and ejection were occurring simultaneously.
EXPERIMENTAL PROCEDURES
Materials—UDP[14C]GlcUA was prepared as described (35). E. coli strain JD424, which expresses the recombinant cellubiuronan synthase from S. pneumoniae strain WU2, has been described (18). Sephacryl S-400, UDP-Glc, UDP-GlcUA, and Streptomyces chromofuscus phospholipase D were from Sigma. Sephacryl S-1000 was from Amersham Biosciences. E. coli membranes containing cellubiuronan synthase were prepared as previously described (13).
Assay of Synthase Activity—Standard cellubiuronan synthase activity determinations were carried out in 100-μl mixtures consisting of 100 mm Hepes (pH 7.5), 10 mm MgCl2, E. coli membranes (3.2 μg of membrane protein), and UDP-Glc and UDP-[14C]GlcUA (0.02 μCi) at the indicated concentrations. The reactions were incubated at 35 °C for 12 min and terminated with 5 μl of 12.5 m acetic acid. The reaction components were separated by ascending paper chromatography on Whatman 3MM paper with a solvent mixture of butanol/acetic acid/water (44:16:40). The chromatograms were cut into 1-cm strips, and polysaccharide (origin product) and oligosaccharide-lipid (migrating slightly faster than Glc) were quantified by liquid scintillation counting. Enzyme activity was calculated as nmol of hexuronic acid incorporated/mg of protein.
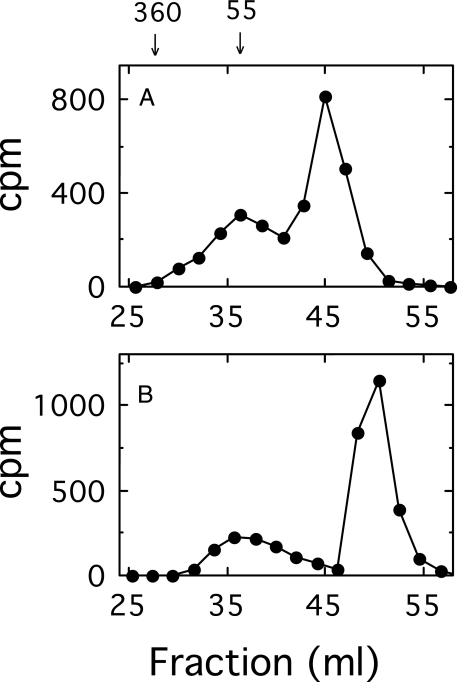
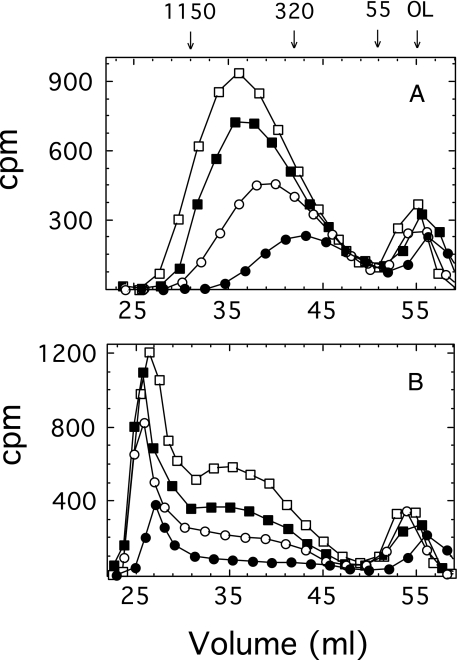
Separation of Oligosaccharide- and Polysaccharide-Lipid by Gel Filtration—A [14C]glucuronyl-labeled mixture of oligosaccharide-lipid and polysaccharide-lipid was synthesized in a 2-ml reaction mixture containing 100 mm Hepes (pH 7.5), 10 mm MgCl2, 2 μm UDP-[14C]GlcUA (106 cpm), 1 mm UDP-Glc, and E. coli membranes (0.5 mg of protein). The reaction was incubated for 25 min at 35 °C and terminated in an ice-water bath. The membranes were washed, and the products were solubilized as described below. Gel filtration chromatography was carried out on a 1.4 × 38-cm column of Sephacryl S-400 irrigated at a flow rate of 20 ml/h with a solution consisting of 10 mm Tris (pH 8.5), 0.1 m NaCl, 0.02% sodium azide, and either 0.1% Nonidet P-40 or 10 mm sodium deoxycholate. Column fractions of 1 ml were collected, and aliquots were assayed for radioactivity by scintillation counting. The polysaccharide synthesized under these conditions was of relatively low molecular weight, and upon gel filtration in a solution containing Nonidet P-40, it was incompletely separated from the detergent micelle region containing the labeled oligosaccharide-lipid (Fig. 1A). By substituting Nonidet P-40 in the column buffer with deoxycholate, which forms much smaller detergent micelles, complete separation was obtained (Fig. 1B). The deoxycholate buffer has greatly facilitated subsequent column analysis. The polysaccharide eluted in a position consistent with a water-soluble glycan with a molecular mass of 55 kDa, and the oligosaccharide-lipid migrated in the detergent micelle elution volume. The relatively small size of the polysaccharide is in part a consequence of the excessive amount of enzyme employed in this experiment and is consistent with a rapid equilibrium ordered mechanism (see “Discussion”).
FIGURE 1.
Effect of detergents on the separation of polysaccharide-lipid from the oligosaccharide-lipid-micelle complex by gel filtration chromatography on Sephacryl S-400. E. coli membranes (0.5 mg of protein) containing cellubiuronan synthase were incubated for 25 min in a 2-ml reaction mixture containing 2 μm UDP-[14C]GlcUA (106 cpm), 1 mm UDP-Glc, 50 mm Hepes, pH 7.5, and 10 mm MgCl2 at 35 °C. The membranes were washed and solubilized, and the products were analyzed by chromatography on Sephacryl S-400, as described under “Experimental Procedures.” The detergents contained in the column elution buffers were 0.1% Nonidet P-40 (A) and 10 mm sodium deoxycholate (B). The numbers above the graph represent cellubiuronan polysaccharide size standards in kDa.
Analytical Methods—Oligosaccharides were released from the oligosaccharide-lipid by digestion with phospholipase D (10 units/100 μl) for 1 h at 30 °C in a mixture containing 50 mm Tris (pH 8), 10 mm CaCl2, and 0.2% Nonidet P-40. The digests were heated at 100 °C for 5 min, and the precipitate was removed by centrifugation at 10,000 × g for 5 min. The liberated oligosaccharides were fractionated by ion exchange chromatography on 0.6 × 2.5 cm columns of DEAE-cellulose, eluted at a flow rate of 0.4 ml/min with a two-step ammonium acetate gradient, as previously described (13). Protein concentration was determined with a Coomassie Brilliant Blue G dye reagent (19) using bovine serum albumin as the protein standard. Type 3 polysaccharide molecular weight sizing standards were prepared as described (35).
Pulse-Chase Analysis by Gel Filtration Chromatography—Polysaccharide- and oligosaccharide-lipid products were synthesized under the indicated reaction conditions. The reactions were terminated in an ice-water bath. The membranes were collected by centrifugation at 100,000 × g for 30 min, and the pellets were suspended in 2.5 ml of a wash buffer consisting of 100 mm Hepes (pH 7.5) and 10% glycerol. The washed membranes were collected by centrifugation, as above. The membrane pellet was washed two more times as described above. Solubilization of the labeled products was carried out by first suspending the pellets in 200 μl of 5 mm EDTA (pH 7.5). The suspension was heated at 100 °C for 4 min followed by the addition of 0.25% Nonidet P-40. Gel filtration chromatography on 1.4 × 38-cm columns of Sephacryl S-400 and S-1000 was carried out as described above. The radioactivity in the fractions under the chased and unchased peaks was tabulated to determine the level of engaged and ejected polysaccharide. The turnover time was calculated by dividing the steady state quantity of engaged polysaccharide by the rate of polysaccharide ejection.
Steady State Fully Processive Activity—Because there is a time lag of greater than 1 min in the transition from the transitory processive state to the fully processive state at 35 °C when both UDP-sugars are present at 10 μm concentrations, polysaccharide formation activity in the first 60 s under these reactions conditions is a measure of the level of pre-existing synthase in the fully processive state. Pre-engaged polysaccharide-synthase complex was prepared as a function of the concentration of UDP-GlcUA and then assayed for fully processive activity in reaction mixtures containing both UDP-sugars at 10 μm concentrations as described. A control was run with membranes preincubated in the absence of UDP-GlcUA, and this value was subtracted from all experimental values.
RESULTS
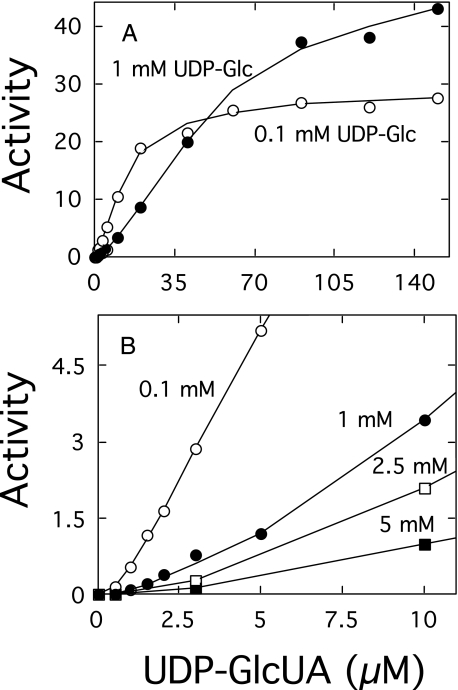
Effect of the Concentration of UDP-sugars on the Rate of Polymerization—The polymerization reaction catalyzed by cellubiuronan synthase proceeds at maximum rates when both UDP-sugars are above 100 μm concentrations (8). Studies in this laboratory have shown, however, that the cellular levels of UDP-Glc in S. pneumoniae are much higher than UDP-GlcUA (15) and that cellubiuronan synthase is able to synthesize high molecular weight polysaccharide at low concentrations of UDP-GlcUA (35). The effect of the concentration of UDP-GlcUA on the rate of the polymerization reaction catalyzed by recombinant cellubiuronan synthase in isolated E. coli membranes at several fixed concentrations of UDP-Glc is shown in Fig. 2. At lower concentrations of UDP-Glc, the reaction rates demonstrated a slight sigmoidal tendency (Fig. 2A). Raising the concentration of UDP-Glc enhanced the sigmoidal-like character of the polymer formation rate curves, indicative of a positive cooperativity (Fig. 2B). These results are quite similar to the effect of increasing temperature, which also resulted in an enhancement in the concavity of the polymer formation rate curves that was coincident with a reduced transition of the cellubiuronan synthase to the fully processive state (13).
FIGURE 2.
Effect of UDP-sugar concentrations on rate of polymer formation. E. coli membranes (3.5μg of protein) containing cellubiuronan synthase were incubated for 12 min at 35 °C in 100-μl standard reaction mixtures containing the indicated concentrations of UDP-GlcUA and 0.1 (○) or 1 mm (•) UDP-Glc (A) and 0.1 (○), 1 (•), 2.5 (□), or 5 mm (▪) UDP-Glc (B). Products were analyzed by paper chromatography as described under “Experimental Procedures.” One unit of activity is defined as the incorporation of 1 nmol of hexose/min/mg of protein. Additional points at higher UDP-GlcUA concentrations are not included in B.
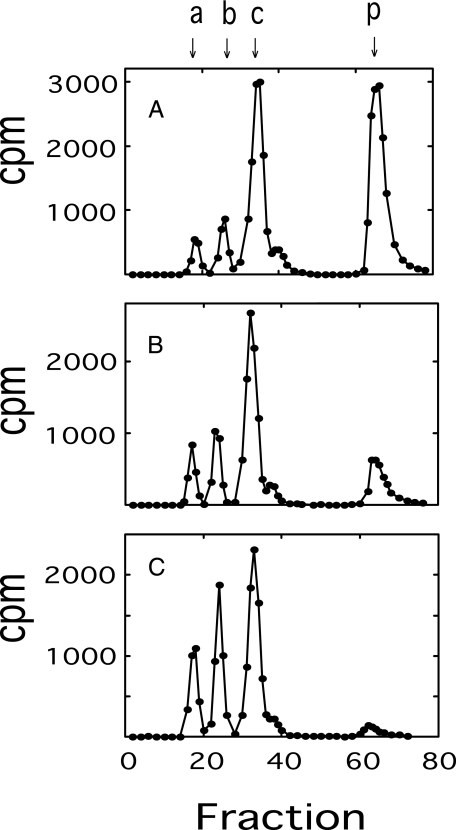
The effects of temperature and UDP-Glc on transition coupling efficiency can be readily distinguished by examining the oligomer size pattern of oligosaccharide-lipid pools synthesized under different temperatures and UDP-Glc concentrations. It was previously shown that the decreased transition efficiency brought about by increasing temperature was accompanied by an elevation in the rate of dissociation of oligosaccharide-lipid during the transitory polymerization state, resulting in a marked increase in the smaller oligomers at higher temperatures (13). As shown in Fig. 3, there was a very significant reduction in polysaccharide formation as the concentration of UDP-Glc was increased, but only a slight effect on the oligomer size pattern was observed. The decreased transition efficiency caused by elevated UDP-Glc appears to be more specifically related to the increased rate of polysaccharide chain ejection that takes place as the concentration of UDP-Glc increases (14). Because the rate of chain ejection was significant with respect to the rate of chain extension, the length of cellubiuronic acid chains was further investigated as described below.
FIGURE 3.
Effect of UDP-Glc concentration on oligomer size of oligosaccharide-lipid products. Reaction mixtures containing 1 μm UDP-[14C]GlcUA (5 × 105 cpm) and 0.1 (A), 1 (B), and 5 mm (C) UDP-Glc were scaled up 10-fold with twice the concentration of E. coli membranes as the standard reaction mixture. The products were washed, solubilized, released by phospholipase D, and analyzed by DEAE-cellulose column chromatography, as described under “Experimental Procedures.” Columns were eluted first with a linear gradient from 0 to 0.3 m ammonium acetate, followed by a 20-ml linear gradient from 0.3 to 1.5 m ammonium acetate. Column fractions of 0.6 ml were collected, and 0.2-ml aliquots were assayed for radioactivity. Cellubiuronan di-, tetra-, hexa-, and polysaccharides are indicated by arrows at a, b, c, and p, respectively.
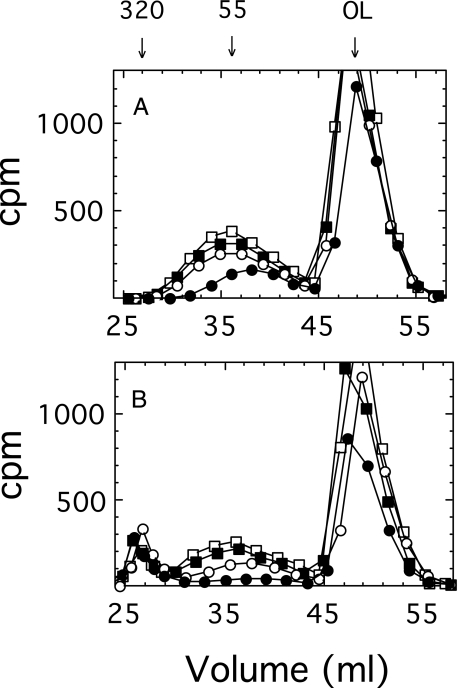
Effect of the Concentration of UDP-GlcUA on Polysaccharide Formation—The size of the growing polysaccharide synthesized with isolated membranes in reaction mixtures containing 1 mm UDP-Glc and 1 μm UDP-GlcUA was monitored on a Sephacryl S-400 column (Fig. 4A). The polysaccharide chains approached a maximum size of ∼50 kDa in less than 12 min and thereafter accumulated without any increase in length. Since the chains remain tethered to the membrane by a lipid primer at their reducing ends following ejection from the synthase (17), there was no direct method to determine the amount of ejected polysaccharide versus that still engaged with the synthase. Therefore, the engaged polysaccharide was chased into high molecular weight product by incubating the membranes for an additional 10 min with a high concentration (0.5 mm) of both UDP-sugars. The polysaccharide engaged with the synthase was indicated by the quantity chased into high molecular weight product (Fig. 4B). The ejected (unchasable) polysaccharide increased in quantity over time, whereas there was a corresponding decrease in the percentage of engaged (chased) polymer. A high percentage of incorporated radioisotope was in the oligosaccharide-lipid product that migrated near the included volume, indicating that much of the synthase remained in the transitory processive state and did not transition into the fully processive state at 1 μm UDP-GlcUA.
FIGURE 4.
Gel filtration profiles of products formed in a reaction containing 1 μm UDP-GlcUA. A, membranes (70 μg of protein) were pulse-labeled in a 2-ml reaction mixture containing 1 mm UDP-Glc, 1 μm UDP-GlcUA (106 cpm), 50 mm Hepes, pH 7.5, and 10 mm MgCl2 at 35 °C. Reactants (0.5 ml) were removed after pulse labeling for 7 (•), 12 (○), 18 (▪), and 24 min (□) and placed in an ice-water bath to terminate the reaction. Carrier membrane (40 μg of protein) was added, and the membranes were centrifuged and washed twice as described under “Experimental Procedures.” One-half of the membranes were saved on ice. B, the remaining pulse-labeled products were chased by incubating the membranes in 200-μl reaction mixtures containing 0.5 mm UDP-Glc, 0.5 mm UDP-GlcUA, 50 mm Hepes, pH 7.5, and 10 mm MgCl2, for 10 min at 35 °C. Carrier membrane (20 μg of protein) was added to all samples, and they were washed once as above. Pulse (A) and chase (B) products were solubilized and analyzed by chromatography on Sephacryl S-400 as described under “Experimental Procedures.” The numbers above the graph represent cellubiuronan polysaccharide size standards in kDa. OL, the elution position of the oligosaccharide-lipid micelle.
In a similar reaction consisting of 6 μm UDP-GlcUA and 1 mm UDP-Glc, the reaction dynamics were markedly different. As shown in Fig. 5A, the steady state polysaccharide was much larger in size (∼500 kDa), and a column of Sephacryl S-1000 was needed to adequately separate the chased and unchasable fractions (Fig. 5B). Approximately 15 min was required to reach maximum chain size, as seen by the column elution position in Fig. 5A, from which point in time the polysaccharide increased in quantity only. The percentage of product that accumulated as oligosaccharide-lipid under these reaction conditions was greatly reduced in comparison with Fig. 4, indicating that the steady state was shifted, with most of the synthase having transitioned into the fully processive state.
FIGURE 5.
Gel filtration profiles of products formed in a 6 μm UDP-GlcUA steady state reaction. Membranes were labeled in a format identical to that in Fig. 4, except the initial reaction contained 7 μm UDP-GlcUA, and the pulse reaction times were for 5 (•), 10 (○), 15 (▪), and 20 min (□). The solubilized pulse (A) and chase (B) products were analyzed on a Sephacryl S-1000 column. Since a steady state was not reached until ∼10% of the reactants had been utilized, the steady state concentration of UDP-GlcUA would have been ∼6 μm.
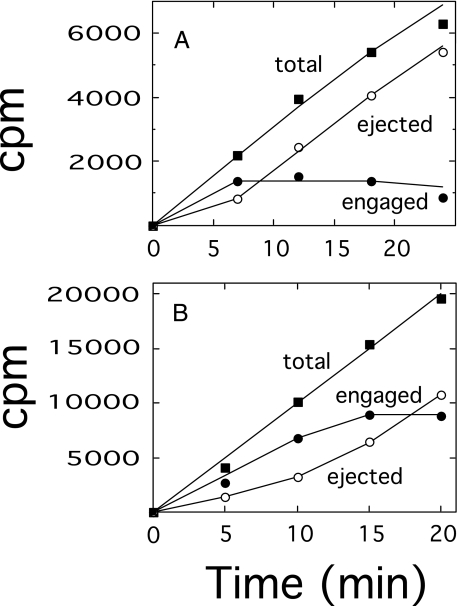
These results demonstrated that chain length modulation proceeded by a steady state assembly process in which polysaccharide was being ejected from the synthase at approximately the same rate as chain initiation. This effect is more clearly shown by plotting the rate of accumulation of engaged and ejected polysaccharide in the reactions depicted above. In a replot of the data obtained at 1 μm UDP-GlcUA, as shown above in Fig. 4, the amount of engaged polysaccharide leveled off at ∼1360 cpm after 7 min (Fig. 6A). The steady state ejection rate was ∼280 cpm/min between 7 and 24 min, indicating a turnover time of ∼5 min (Fig. 6A). It should be emphasized that minimal amounts of enzyme were included in the reaction mixtures in order to maintain a linear rate of polymer synthesis during the course of the reaction and to minimize any decrease in the concentration of UDP-GlcUA over the reaction time period. This objective was particularly difficult to fully achieve in the reaction of Fig. 4, consisting of 1 μm UDP-GlcUA, and the slight reduction in rates after 20 min is probably a reflection of a slight reduction in the concentration of UDP-GlcUA below 1 μm. A replot of the data obtained at 6 μm UDP-GlcUA, as shown in Fig. 5, revealed a steady state engagement of ∼8900 cpm and a steady state ejection rate of ∼880 cpm/min that was attained after 15 min (Fig. 6B). These results yielded an approximate turnover time of 10 min, which was too lengthy to allow for significant enzyme recycling. In a similar experiment at 4 μm UDP-GlcUA, the length of the polysaccharide chains and the rate of turnover were intermediate to those at 1 and 6 μm (data not shown).
FIGURE 6.
Steady state polysaccharide formation at 1 and 6 μm UDP-GlcUA. Engaged polysaccharide, as represented by the chased radioactivity (•); ejected polysaccharide, as represented by the unchased radioactivity (○); and the total of both chased and unchased polymer (▪) were determined as described under “Experimental Procedures” and plotted as a function of time. A, data from Fig. 4. B, data from Fig. 5.
These results indicate a kinetic model wherein relative polysaccharide chain length can be predicted based on the rate of the reaction compounded by the time of chain engagement with the synthase binding site at any fixed concentration of UDP-GlcUA. Since the reaction rates shown in Fig. 2 do not reflect the steady state amount of enzyme in the fully processive state, the proportion of enzyme in each state as a function of the UDP-GlcUA concentration was determined at both 0.1 and 1 mm UDP-Glc (Fig. 7). As expected, the synthase transitioned into the fully processive mode much more readily at the lower concentration of UDP-Glc. The relatively low level of synthase that transitions into the fully processive state in the presence of high concentrations of UDP-Glc may reflect a partial inactivation that occurs at highly imbalanced concentrations of the cognate UDP-sugars.3
FIGURE 7.
Steady state proportion of synthase in the fully processive state. Washed E. coli membranes (66 μg of protein) were preloaded with polysaccharide by incubating for 6 min at 35 °C in 2-ml reaction mixtures containing 100 mm Hepes, pH 7.5, 10 mm MgCl2, and either 0.1 (○) or 1 mm UDP-Glc (•), at the indicated concentrations of UDP-GlcUA. The reactions were terminated in an ice-water bath, and the membranes were washed twice as described in the legend to Fig. 4. The membranes were suspended in 0.1 ml of ice-cold 100 mm Hepes, pH 7.5. As explained under “Experimental Procedures,” the amount of fully processive activity was then determined by incubating the preloaded membranes (30 μl) in 0.2-ml reaction mixtures containing 10 μm UDP-[14C]GlcUA (105 cpm), 10 μm UDP-Glc, 100 mm Hepes, pH 7.5, and 10 mm MgCl2, previously brought to 35 °C. The reactions were incubated for 60 s at 35 °C, terminated by the addition of 5 μl of 12.5 m acetic acid, and the products were separated by paper chromatography. The formation of polysaccharide (origin product) is indicated as the percentage of fully processive activity of synthase in the membranes preincubated at the indicated concentration of UDP-GlcUA.
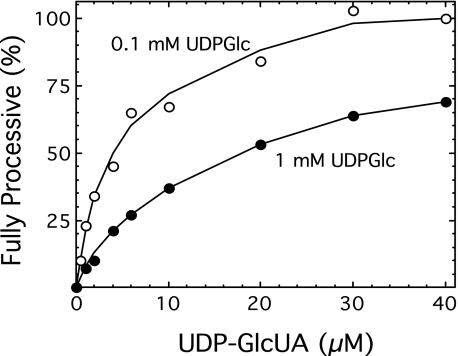
The fully processive activities, based on the proportion of enzyme in the fully processive state at 1 mm UDP-Glc, were then calculated as shown in Table 1. These values, in conjunction with the length of time that the growing chain was engaged with the carbohydrate substrate recognition site, were used in the kinetic model to generate the relative polysaccharide chain lengths. The relative chain lengths were normalized to the chemically determined chain length of 320 residues at 1 μm UDP-GlcUA to generate a series of predicted chain lengths, which were in good agreement with the chemically determined chain lengths at the respective UDP-GlcUA concentrations (Table 1).
TABLE 1.
Kinetic model of cellubiuronan polysaccharide chain length regulation by UDP-GlcUA at 1 mm UDP-Glc
|
UDP-GlcUA
|
Total activity
|
Fully processive activity
|
Turnover
timeb
|
Relative polymer
sizec
|
Kinetic model
lengthd
|
Chemically determined
lengthe
|
||
|---|---|---|---|---|---|---|---|---|
| Percentage | Amounta | Relative rate | ||||||
| μm | nmol/min/mg | % | nmol/min/mg | min | residues | residues | ||
| 1 | 0.1 | 8 | 1.25 | 1.0 | 5 | 1.0 | 320 | 320 |
| 2.5 | 0.5 | 15 | 3.33 | 2.7 | (7) | 3.7 | 1195 | 1160 |
| 4 | 0.9 | 21 | 4.29 | 3.4 | 8 | 5.5 | 1755 | 1900 |
| 6 | 1.6 | 27 | 5.93 | 4.7 | 10 | 9.5 | 3034 | 3100 |
| 11.5 | 4.2 | 40 | 10.5 | 8.4 | (12) | 20.2 | 6451 | 6700 |
Fully processive activity was determined by dividing the total activity values extracted from Fig. 2 by the percent of synthase in the fully processive state as determined in Fig. 7.
Values in parenthesis were estimated.
Relative polymer size was calculated by multiplying the relative fully processive rate by the relative turnover time. The latter was obtained by dividing the turnover time by 5.
The kinetic model chain length at 1 μm UDP-GlcUA was accorded the identical value of 320 residues as had been previously determined by a chemical method, and the remaining kinetic model values were calculated by normalizing the relative polymer sizes with the 1 μm value.
Chemically determined chain lengths were from the accompanying paper (35).
Results obtained at 0.1 mm UDP-Glc also appear to be consistent with the kinetic model. Cellubiuronan synthesized in a reaction containing 1 μm UDP-GlcUA and 0.1 mm UDP-Glc was chemically determined to have a chain length of 1500 sugar residues (data not shown). This approximate 5-fold increase in chain length (compare with 320 residues at 1 μm UDP-GlcUA and 1 mm UDP-Glc in Table 1) is in agreement with a slightly greater than doubling of both the relative fully processive activity and turnover time in the reaction at 0.1 mm UDP-Glc. The relationship between chain size and UDP-sugar concentrations was highly reproducible between duplicate experiments and with different enzyme preparations, as verified by gel filtration chromatography and depolymerase chain length analysis (35).
As shown in Table 1, the rate of the fully processive reaction is relatively linear at low concentrations of UDP-GlcUA, consistent with hyperbolic kinetics. This is in contrast to the sigmoidal character at 1 mm UDP-Glc, as shown in Fig. 2, where no correction was made for the percentage of synthase in the transitory state. From an allosteric viewpoint, the cooperativity demonstrated in Fig. 2 appears to be the result of an enhancement in the amount of synthase in the fully processive state rather than an effect on some other kinetic parameter of the polymerization reaction. These results are in agreement with the previously indicated catalytic restructuring that occurs in conjunction with the formation of a tightly binding enzymepolysaccharide complex (13).
DISCUSSION
There is a correlation between the length of the polysaccharide chain synthesized by cellubiuronan synthase and the concentration of UDP-GlcUA present in the polymerization reaction (35). The validity of these findings is shown in greater detail here in the steady state kinetics of polysaccharide production, which provide a fuller demonstration of how cellubiuronan synthase functions as a concentration-dependent timing device to regulate the final chain length of the polysaccharide product. The data suggest that the timing mechanism is integrated within the peptide matrix such that it is chemically sensitive to the concentration of the UDP-sugars and physically sensitive to the temperature (13). Hence, the temperature- and concentration-dependent processive assembly of S. pneumoniae capsular polysaccharide may offer a new perspective on the study of processive β-glycan polymerization reactions. In particular, the mechanical aspects of this processive assembly (35) need to be more fully considered, as has been emphasized for other complex biological reactions (20).
Mechanisms that would specifically modulate chain length by regulating the processive assembly of cellubiuronan and related polysaccharides, such as hyaluronan, cellulose, chitin, and others, are only poorly understood (21). Control of polymer size by substrate concentration has been suggested for several polysaccharide systems, but data supporting this hypothesis is limited (15). It would logically be expected for most polysaccharide polymerization reactions that the rate of the reaction would increase as the nucleotide sugar donor concentration increases, and hence a simple correlation of chain size with the substrate concentration of an in vitro reaction is not, in and of itself, sufficient evidence that the chain length is regulated by the donor concentration. Solid evidence for chain length regulation and a more complete explanation of the nucleotide sugar substrate concentration-dependent relationship of the assembly of cellubiuronan chains required the clear demonstration of a steady state production of a unique chain size at different UDP-sugar concentrations, as has been shown here. These results have allowed the development of a kinetic model wherein the chain length was predicted based on the rate of synthesis compounded by the length of time that the growing chain remains engaged at the carbohydrate acceptor substrate-binding site.
Cellubiuronan synthase is similar in protein homology to a number of polysaccharide synthases, including hyaluronan, cellulose, chitin, and several other synthases (11, 18), and is thought to contain a single catalytic center that could alternately bind both UDP-sugars (22). It was proposed that a single nucleotide sugar center could catalyze the synthesis of polymer consisting of a repeating disaccharide if the addition of each sugar served to fine tune the affinity of the donor site for the succeeding nucleotide sugar (22). Studies with the cellubiuronic acid synthase have shown that the bound oligosaccharide substrate apparently acts as a cofactor in determining the affinity of nucleotide sugar donor substrates and that in the absence of bound carbohydrate, the enzyme demonstrates deceased fidelity in recognizing the appropriate acceptor substrate (13). Possibly, these changes in affinity reflect a related restructuring similar to that of several glycosyltransferases (23–26), where the ordered binding of metal ion and nucleotide sugar to the donor domain induces a peptide conformational change, which in turn creates a binding site in the acceptor domain (27, 28).
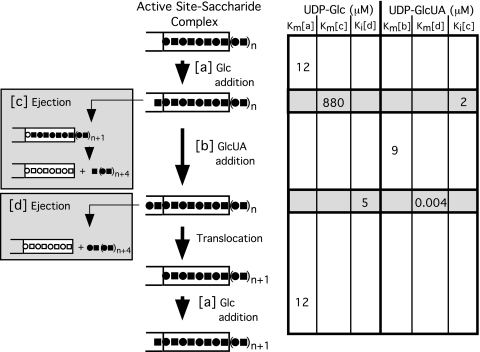
An induction scheme for the cyclical disaccharide extension of type 3 polysaccharide (Fig. 8) provides more detail and enlarges a previous more conceptual model (13). Each catalytic cycle entails the alternate association of the synthase with UDP-Glc and UDP-GlcUA, the formation of the glycosidic linkages of the respective sugars, and the release, translocation, and reattachment of the elongating chain at the synthase-carbohydrate recognition site. The focus of the model in Fig. 8 is on the reciprocal interplay between the nucleotide sugar binding pocket, which catalyzes the transfer of Glc (a) or GlcUA (b) and the saccharide complex, which cyclically translocates the growing chain following the addition of both sugars, and ejects the chain (c) when limiting levels of UDP-GlcUA are present. Cellubiuronan synthase requires a minimum of a tetrauronosyl oligomer for recognition by the carbohydrate substrate binding site (13). Apparently, if the translocation process is energized prior to completion of a new disaccharide unit in the catalytic pocket, the chain is ejected (c) because it has shifted too far out of alignment with the carbohydrate recognition site. It is not definitively known whether translocation occurs after Glc or GlcUA addition; however, the 50-fold greater Vmax of ejection energized by UDP-Glc compared with UDP-GlcUA (14) would be consistent with the most frequent misalignment occurring with polysaccharide movement following Glc rather than GlcUA addition. A rare ejection (d) event also occurs following the GlcUA addition but may be of minimal physiological significance, as discussed below.
FIGURE 8.
Reciprocal sugar addition and disaccharide extension by sequential induction of the fully processive reaction. Kinetic constants are shown on the right for UDP-Glc and UDP-GlcUA in line with the reactions shown to the left. The apparent Km(a) value for UDP-Glc and apparent Km(b) value for UDP-GlcUA are from polymerization studies with S. pneumoniae membranes (8). The apparent Km(c) and Ki(d) values for UDP-Glc and the apparent Km(d) and Ki(c) values for UDP-GlcUA are from ejection studies with S. pneumoniae membranes (14). Residues of Glc (▪) and GlcUA (•) and unfilled binding sites in the carbohydrate acceptor recognition site for Glc (□) and GlcUA (○) are indicated.
We previously suggested that the polymerization reaction is rapid equilibrium-ordered since translocation of the polysaccharide chain would presumably be slow in comparison with the association and dissociation of the nucleotide sugar precursors. This suggestion is supported by the relatively large activation energy for cellubiuronan synthesis in S. pneumoniae (14), which was similar to that for hyaluronan synthesis (29), and would be consistent with the slowness of formation of similar polymers (9). A rapid equilibrium-ordered mechanism is also consistent with the effect of excessive levels of enzyme (see Fig. 1), which effectively lowered the substrate concentrations, resulting in a shorter than expected polysaccharide chain length in a reaction containing 2 μm UDP-GlcUA.
Since Km and Ks values are equivalent in rapid equilibrium reactions, the apparent Km(a) and Km(b) values of 12 and 9 μm for UDP-Glc and UDP-GlcUA, respectively, should reflect their affinities for the synthase in the fully processive reaction (Fig. 8). This correlation is corroborated by the Ki values, a direct measure of affinity, in inhibiting the ejection actuated by the alternate nucleotide sugar. UDP-Glc inhibits polysaccharide chain ejection by UDP-GlcUA with a Ki(d) of 5 μm, similar to its Km(a) of 12 μm in the processive reaction. UDP-GlcUA inhibits ejection by UDP-Glc with a Ki(c) of 2 μm, similar to its Km(b) of 9 μm in the processive reaction. Inhibition of the ejection event apparently occurs as a consequence of the glycosidic addition by the alternate nucleotide sugar before ejection takes place, as described in more detail below.
The rate of chain ejection (c) actuated by UDP-Glc is exceedingly slow, being many orders of magnitude slower than the rates of association and dissociation of UDP-Glc with the enzyme (Fig. 8). Thus, it would be expected that in the absence of sufficient UDP-GlcUA to continue chain extension, the addition of Glc would occur well in advance of ejection on a catalytic time scale. The apparent Km(c) value of 880 μm determined for the UDP-Glc ejection in S. pneumoniae membranes in the absence of UDP-GlcUA (14) presumably reflects the decreased affinity of the synthase for UDP-Glc following the addition of glucose to the growing chain. Thus, the presence of the glucosyl chain extension in the catalytic pocket appears to decrease the affinity of the synthase for UDP-Glc by 2 orders of magnitude. In the normal reaction sequence occurring when both UDP-sugars are present, the subsequent addition of GlcUA followed by translocation apparently restores the affinity for UDP-Glc to its original level. These observations are in agreement with the sequential induction process wherein the terminal sugar could serve to fine tune the affinity of the synthase for the appropriate substrate in the catalytic sequence by a reciprocal enhancement of the affinity of the synthase for the complementary UDP-sugar.
A very slow ejection (d) occurs when UDP-GlcUA is present in the absence of UDP-Glc (Fig. 8). Since UDP-Glc is a precursor to UDP-GlcUA and is virtually always present in the cell at higher concentrations than UDP-GlcUA and additionally because of the sluggishness of this ejection event, the ejection by UDP-GlcUA is probably of no physiological significance. However, from a mechanistic point of view, it may be of some interest that under these conditions, UDP-GlcUA was determined to have an apparent Km(d) of 4 nm in the ejection reaction with S. pneumoniae membranes (14). In the absence of more definitive information concerning the mechanics of the translocation step, it can only be speculated as to the significance of this large apparent increase in affinity of UDP-GlcUA for the synthase nucleotide sugar-binding site. Taking a cue from the above effect with UDP-Glc, the data may indicate that a glucuronyl-glucosyl extension into the catalytic pocket dramatically alters the affinity for UDP-GlcUA, albeit in a very unusual manner. It should also be noted that a second binding site for UDP-GlcUA cannot be ruled out. Whatever the final explanation, the apparent change in affinity by 3 orders of magnitude for UDP-GlcUA is consistent with a sequential induction process. Following the shift of the polysaccharide chain during translocation, the affinity of the synthase would revert to the initial conditions allowing the entry of UDP-Glc to initiate another cycle of sugar addition.
Recent studies of several glycosyltransferases in which binding of the nucleotide sugar donor substrate creates in turn a carbohydrate acceptor-binding site (30), may shed some light on the transduction potential of processive glycosyltransferases. It can be speculated that following sugar addition to the growing chain during processive polysaccharide assembly, the extension of the distal chain sugar into the catalytic pocket may create bonding and distortional forces to which subsequent incoming nucleotide sugars might be subjected. In turn, the binding of a nucleotide sugar may exert distortional forces on the interaction between the polysaccharide chain and the carbohydrate acceptor-binding site. A variety of induction schemes along these lines may be involved in the many diverse types of processive polysaccharide assemblies.
In view of the large diversity and adaptability of carbohydrate binding domains of glycosyltransferases and hydrolases, there is an increasing awareness of their potential in biological recognition and signaling phenomena (31). This capacity has been demonstrated by the Golgi β-galactosyltransferase to also function as a lectin-like recognition component in a number of cellular interactions (32, 33). A related phenomenon may be the coordination of biosynthesis, termination, and export of the lipopolysaccharide O-antigen following its binding by the nucleotide-binding domain (Wzt) component of the ABC transporter in E. coli (34). The versatility of the catalytic center of the cellubiuronan synthase is apparent in that it initiates oligomer formation on the internal membrane surface and then undergoes a restructuring to provide for the extrusion of the growing chain on the external membrane surface while monitoring the internal nucleotide sugar concentrations that modulate the polymerization in determining the external capsule chain length.
This work was supported, in whole or in part, by National Institutes of Health, NIGMS, Grant R01 GM53017.
Footnotes
The abbreviation used is: GlcUA, glucuronic acid.
R. T. Cartee, Y. Sutanto, and J. Yother, manuscript in preparation.
References
- 1.Kim, J. O., and Weiser, J. N. (1998) J. Infect. Dis. 177 368–377 [DOI] [PubMed] [Google Scholar]
- 2.Magee, A. D., and Yother, J. (2001) Infect. Immun. 69 3755–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wessels, M. R., and Bronze, M. S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 12238–12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telsor, A., Robinson, H. C., and Dorfman, A. (1966) Arch. Biochem. Biophys. 116 458–465 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell, D., and Hardingham, T. (1982) Biochem. J. 202 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, B. R., Cuthbertson, L., and Whitfield, C. (2004) J. Biol. Chem. 279 35709–35718 [DOI] [PubMed] [Google Scholar]
- 7.Carter, J. A., Blondel, C. J., Zaldivar, M., Alvarez, S. A., Marolda, D. L., Valvano, M. A., and Contreras, I. (2007) Microbiology 153 3499–3507 [DOI] [PubMed] [Google Scholar]
- 8.Cartee, R. T., Forsee, W. T., and Yother, J. (2000) J. Biol. Chem. 275 3907–3914 [DOI] [PubMed] [Google Scholar]
- 9.Lidholt, K., Riesenfeld, J., Jacobsson, K.-G., Feingold, D. S., and Lindahl, U. (1988) Biochem. J. 254 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena, I. M., Brown, R. M. J., Fevere, M., Geremia, R. A., and Henrissat, B. (1995) J. Bacteriol. 177 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keenleyside, W. J., and Whitfield, C. (1996) J. Biol. Chem. 271 28581–28592 [DOI] [PubMed] [Google Scholar]
- 12.Lairson, L. L., Henrissat, B., Davies, G. J., and Withers, S. G. (2008) Annu. Rev. Biochem. 77 521–555 [DOI] [PubMed] [Google Scholar]
- 13.Forsee, W. T., Cartee, R. T., and Yother, J. (2006) J. Biol. Chem. 281 6283–6289 [DOI] [PubMed] [Google Scholar]
- 14.Forsee, W. T., Cartee, R. T., and Yother, J. (2000) J. Biol. Chem. 275 25972–25978 [DOI] [PubMed] [Google Scholar]
- 15.Ventura, C. L., Cartee, R. T., Forsee, W. T., and Yother, J. (2006) Mol. Microbiol. 61 723–733 [DOI] [PubMed] [Google Scholar]
- 16.Cartee, R. T., Forsee, W. T., Jensen, J. W., and Yother, J. (2001) J. Biol. Chem. 276 48831–48839 [DOI] [PubMed] [Google Scholar]
- 17.Cartee, R. T., Forsee, W. T., and Yother, J. (2005) J. Bacteriol. 187 4470–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillard, J. P., Vandersea, M. W., and Yother, J. (1995) J. Exp. Med. 181 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 20.Bustamante, C., Chemla, Y. R., Forde, N. R., and Izhaky, D. (2004) Annu. Rev. Biochem. 73 705–748 [DOI] [PubMed] [Google Scholar]
- 21.Whitfield, C. (2006) Annu. Rev. Biochem. 75 39–68 [DOI] [PubMed] [Google Scholar]
- 22.Charnock, S. J., Henrissat, B., and Davies, G. J. (2001) Plant Physiol. 125 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brew, K., Vanaman, T. C., and Hill, R. L. (1968) Proc. Natl. Acad. Sci. U. S. A. 59 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison, J. F., and Ebner, K. E. (1971) J. Biol. Chem. 246 3977–3984 [PubMed] [Google Scholar]
- 25.Kharta, B. S., Herries, D. G., and Brew, K. (1974) Eur. J. Biochem. 44 537–560 [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa, Y., Pegg, W., Paulsen, H., and Schachter, H. (1988) J. Biol. Chem. 263 8270–8281 [PubMed] [Google Scholar]
- 27.Ramakrishnan, B., Balaji, P. V., and Qasba, P. K. (2002) J. Mol. Biol. 318 491–502 [DOI] [PubMed] [Google Scholar]
- 28.Ramakrishnan, B., Ramasamy, V., and Qasba, P. K. (2006) J. Mol. Biol. 357 1619–1633 [DOI] [PubMed] [Google Scholar]
- 29.Pummill, P. E., Achyuthan, A. M., and DeAngelis, P. L. (1998) J. Biol. Chem. 273 4976–4981 [DOI] [PubMed] [Google Scholar]
- 30.Qasba, P. K., Ramakrishnan, B., and Boeggeman, E. (2005) Trends Biochem. Sci. 30 53–62 [DOI] [PubMed] [Google Scholar]
- 31.Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004) Biochem. J. 382 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodeheffer, C., and Shur, B. D. (2002) Biochim. Biophys. Acta 1573 258–270 [DOI] [PubMed] [Google Scholar]
- 33.Ramakrishnan, B., Shah, P. S., and Qasba, P. K. (2001) J. Biol. Chem. 276 37665–37671 [DOI] [PubMed] [Google Scholar]
- 34.Cuthbertson, L., Kimber, M. S., and Whitfield, C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19529–19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsee, W. T., Cartee, R. T., and Yother, J. (2009) J. Biol. Chem. 284 11826–11835 [DOI] [PMC free article] [PubMed] [Google Scholar]