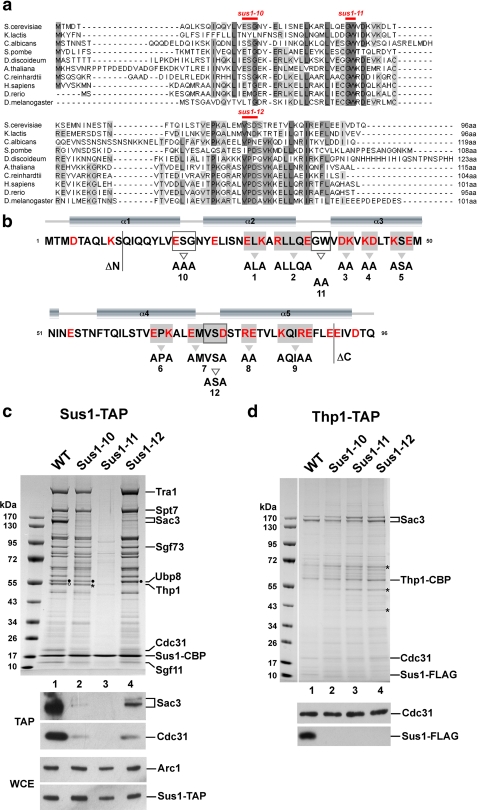
FIGURE 1.
Conserved residues differentially regulate association of Sus1 with SAGA or TREX-2. a, multiple sequence alignment of Sus1 from representative species including fungi, protozoa, plants and metazoa. The alignment was generated with ClustalW2, and conserved residues were shaded using JalView. b, secondary structure prediction for Sus1 was calculated from PsiPred. For schematic depiction of Sus1 mutational strategies, mutant alleles are numbered (sus1ΔN, sus1–1 to sus1–12, sus1ΔC). All Sus1 charged residues are indicated in red. Residues mutated according to the clustered charge to alanine algorithm are shaded in gray, and gray arrowheads denote sequence changes. Open boxes/open arrowheads depict (partially) conserved Sus1 residues/sequence changes with a predicted localization within protein loops. N- and C-terminal truncations are indicated. Positions of sus1–10, sus1–11 and sus1–12 mutations are also indicated in a. c, affinity purification of genomically expressed Sus1-TAP from wild-type (WT), sus1–10, sus1–11, and sus1–12 strains. Eluates were analyzed by SDS-PAGE (4–12% gradient gels, MOPS buffer) and Coomassie staining. Indicated bands were assigned according to their molecular mass (15). Filled circles designate Ubp8 and open circles designate Thp1. Asterisk indicates a contaminant, eEF-1α, which runs slightly below Thp1. Ubp8, Thp1, and eEF-1α were determined by mass spectrometry. Eluates (TAP) were analyzed by Western blotting using anti-Sac3 and anti-Cdc31 antibodies. Sus1-TAP expression levels (WCE) were determined by anti-ProtA detection (ProtA is part of the TAP tag) and normalization with Arc1 (a yeast cytosolic marker protein) levels. d, Thp1-TAP affinity purification from the indicated sus1 mutant allele backgrounds. Indicated Coomassie-stained bands were determined by mass spectrometry. Asterisks label contaminants of the purification (from top to bottom: keratin, Eno2, and Tdh3). FLAG-tagged Sus1 and Cdc31 were immunodetected by anti-FLAG and anti-Cdc31 antibodies, respectively.