Abstract
The TREX enzymes process DNA as the major 3′→5′ exonuclease activity in mammalian cells. TREX2 and TREX1 are members of the DnaQ family of exonucleases and utilize a two metal ion catalytic mechanism of hydrolysis. The structure of the dimeric TREX2 enzyme in complex with single-stranded DNA has revealed binding properties that are distinct from the TREX1 protein. The TREX2 protein undergoes a conformational change in the active site upon DNA binding including ordering of active site residues and a shift of an active site helix. Surprisingly, even when a single monomer binds DNA, both monomers in the dimer undergo the structural rearrangement. From this we have proposed a model for DNA binding and 3′ hydrolysis for the TREX2 dimer. The structure also shows how TREX proteins potentially interact with double-stranded DNA and suggest features that might be involved in strand denaturation to provide a single-stranded substrate for the active site.
INTRODUCTION
The 3′→5′ exonucleases provide an essential function in a variety of DNA metabolic pathways. Traditionally thought of as primarily processing 3′-termini for DNA replication, repair and recombination events, our understanding of their roles has grown to include other metabolic pathways, such as degrading single-stranded DNA (ssDNA) to prevent autoimmune dysfunction (1–4). The TREX2 and homologous TREX1 enzymes provide the major 3′-exonuclease activity observed in mammalian cell extracts (5–7). Recently, mutations in the TREX1 gene have been identified as the underlying cause of the neurological brain disease, Aicardi–Goutieres syndrome and familial chilblain lupus, both diseases having pathologies related to an aberrant immune response characterized by symptoms that include induction of type I interferons and circulating auto-antibodies reactive to single-stranded and double-stranded DNA (dsDNA). One function of TREX1 is to degrade ssDNA derived from processing of aberrant replication intermediates or from endogenous retroelements to prevent their accumulation and activation of cell-intrinsic antiviral defense (4,8). The TREX1 protein has also been identified as the granzyme-A activated exonuclease within the SET complex, an endoplasmic reticulum (ER)-associated complex that contains the SET protein, the endonuclease, NM23-H1 and the base excision repair exonuclease, Ape1 as well as several other proteins (1). The interaction with this complex is specific for the TREX1 protein and not TREX2 (1), leaving the cellular role of TREX2 undefined and suggesting differing biological functions for these two enzymes.
The TREX1 and TREX2 enzymes are Mg2+ ion-dependent deoxyribonucleases belonging to the DnaQ-like exonuclease family. The DnaQ family has been divided into two subfamilies characterized by the presence of four conserved carboxylate residues and a histidine (DEDD-h) or a tyrosine (DEDD-y) positioned in the active site. The side chains of the four conserved carboxylate residues in TREX1 and TREX2 coordinate two Mg2+ ions and the histidine is positioned to deprotonate a water producing a nucleophile to attack the target phosphodiester bond and effect hydrolysis. The TREX1 and TREX2 enzymes are the only known mammalian deoxyribonuclease members of the DEDD-h subfamily and prefer excision of deoxynucleotide substrates over ribonucleotides by about 1000-fold (6). The TREX1 and TREX2 proteins share about 40% sequence identity and not surprisingly similar overall 3D structures (9–11). However, distinct differences between the proteins support the idea of independent cellular functions for these two exonucleases. The TREX1 protein contains an extended C-terminal domain of about 76 amino acids not present in TREX2 that plays a putative role in anchoring TREX1 to the cytosolic side of the ER membrane (12). The TREX1 protein also contains a non-repetitive proline-rich region not present in TREX2 that is thought to play a specific role in protein–protein interactions, such as that with the SET complex (9,10). Additionally, kinetic data for the TREX enzymes reveal about a 10-fold lower efficiency for TREX2 relative to that of TREX1 (6,11,13,14).
Our previous characterization of the TREX2 protein revealed a mobile loop adjacent to the active site provides the major DNA binding element and contributes to the catalytic efficiency of the enzyme (11,15). Mutations of three arginine residues within this loop of TREX2 resulted in a 60-fold decrease exonuclease activity; a result of increased KM with no effect on kcat. We have also shown evidence for cooperative DNA binding with the TREX2 protein and coordinated catalysis between the protomers of the TREX2 dimer (15). Heterodimers of TREX2 containing one wild-type protomer and one catalytically inactive protomer (TREX2wt/H188A) showed a 7-fold decrease in catalytic activity in contrast to a 2-fold lower activity in the DNA binding heterodimer (TREX2wt/R163A-R165A-R167A). The decrease in the TREX2wt/H188A heterodimer indicates that the defective catalysis in one protomer reduces the activity in the opposing protomer.
In order to understand the structural aspects of the TREX2 protein that contribute to the coordinated catalysis and identify a possible mechanism for communication between the active sites, we determined the crystal structure of the TREX2 dimer in complex with ssDNA and Ca2+. The structure revealed several novel aspects of TREX2–DNA interaction including a conformational change in the active site upon binding that is communicated across the dimer interface, as well as a sequential mode of binding DNA and divalent metal ion. Additionally, we have identified a structural motif that may contribute to displacement of dsDNA to provide single-stranded substrate to the active site.
MATERIALS AND METHODS
Protein expression, purification and crystallization
The human TREX2 protein was expressed and purified as previously described (10,11,16). The protein–nucleic acid complexes were crystallized by the hanging drop vapor diffusion technique using TREX2–DNA complex formed by mixing protein with a ssDNA oligonucleotide (GACG) in a molar ratio of 1 : 2 in the presence of 2 mM CaCl2. Two microlitres of protein complex at 5 mg/ml was mixed with equal volume of reservoir solution and suspended on a cover slip above the 200 μl reservoir. The TREX2–DNA complex crystallized using 4.5% PEG 4000, 5% 2-methyl-2,4-pentanediol (MPD), 50 mM HEPES pH 7.2, 200 mM NaCl and 2 mM CaCl2. Crystallization experiments were carried out at 20°C and crystals appeared within 1 week.
X-ray data collection, phasing and refinement
Prior to data collection crystals were soaked in reservoir solution containing 6% PEG 4000, 7% MPD, 50 mM HEPES, pH 7.2, 200 mM NaCl and 2 mM CaCl2 for 3–5 days. Crystals were then mounted in a nylon loop and flash frozen in liquid nitrogen. X-ray data were collected using Cu Kα radiation on a MicroMax 007 generator and a Saturn 92 CCD detector (Rigaku). Intensity data were processed with the program d*TREK (17). Crystals of the TREX2–DNA complex belong to space group P21 with unit cell dimensions a = 55.5 Å, b = 78.1 Å, c = 162.9 Å, α = γ = 90°, β = 90.6° (Table 1).
Table 1.
Crystallographic data and refinement statistics
| Complex | TREX2/ssDNA |
|---|---|
| Space group | P21 |
| Unit cell (Å) | a = 55.51, b = 78.08, c = 162.88; α = 90°, β = 90.6°, γ = 90° |
| Molecules (a.u.) | 6 |
| Wavelength (Å) | 1.54 |
| Resolution (Å) | 20–3.1 |
| Completeness (%) | 94.4 (92.4) |
| Rmerge (%)a | 14.5 (36.1) |
| Mean (I/σ) | 3.3 (2.1) |
| Average redundancy | 2.6 (2.3) |
| R-factor (%)b | 25.4 |
| Rfree (%) | 30.0 |
| Average B-factor protein (Å2) | 34.5 |
| Average B-factor nucleotide (Å2) | 44.6 |
| Average B-factor water (Å2) | 19.4 |
| RMSD bond lengths (Å) | 0.015 |
| RMSD bond angles (degree) | 2.92 |
aRmerge = Σ|I − <I>|/ΣI, where I is the observed intensity and <I> is the average intensity.
bR-factor = Σ||FO| − |FC||/Σ|FO|. Rfree is the same as R, but calculated with 10% of the reflections that were never used in crystallographic refinement.
Statistics for outer resolution shell are given in parentheses (3.1–3.2 Å).
Phases for the data were obtained by molecular replacement using the program PHASER (18) and a monomer of the hTREX2 apo protein (pdbid: 1Y97) (11) as a search model. The TREX2–ssDNA–Ca2+ model was built using the program COOT (19). The structure was refined without restraints of non-crystallographic symmetry (NCS) using the programs CNS and Refmac5 (20,21), and the validity of the refined structure confirmed by simulated annealing-omit procedures (20). The change in the free R-factor was monitored at each step in refinement, as well as the inspection of stereochemical parameters with the programs Molprobity (22) and ERRAT (23). The model of the complex converged with a final R-factor of 25.5% (Rfree = 30.0%) using all observed X-ray data measurements in the resolution range 20–3.0 Å. A Ramachandran plot shows that >90% of all residues in the models have Φ and ψ angles in the most preferred regions. All figures of molecular structures were created using the program PyMOL. Atomic coordinates for the TREX2–ssDNA–Ca2+ complex have been deposited in the Protein Data Bank.
RESULTS AND DISCUSSION
Structure of TREX2–DNA complex
We crystallized the human TREX2 protein in complex with ssDNA and Ca2+. The structure was determined by molecular replacement using the unliganded TREX2 protein as a search model (Table 1). The correct solution provided an initial R-factor of about 46% after a round of rigid body refinement. The asymmetric unit of the crystals contains three dimers of the TREX2 protein. Electron density maps revealed that one of the TREX2 dimers has DNA bound in both active sites while the other two have DNA bound in only a single active site, indicating a sequential binding mechanism for TREX2 (Figure 1). A superposition of a TREX2 monomer with bound DNA onto each of the holo monomers in the asymmetric unit confirms that there is room to accommodate the oligonucleotide and that the lack of binding is not due to crystal packing or other steric hindrances. The details of the dimeric nature of the TREX2 protein in the absence of DNA have been described previously (11) and are similar to our current structure with the exceptions described below.
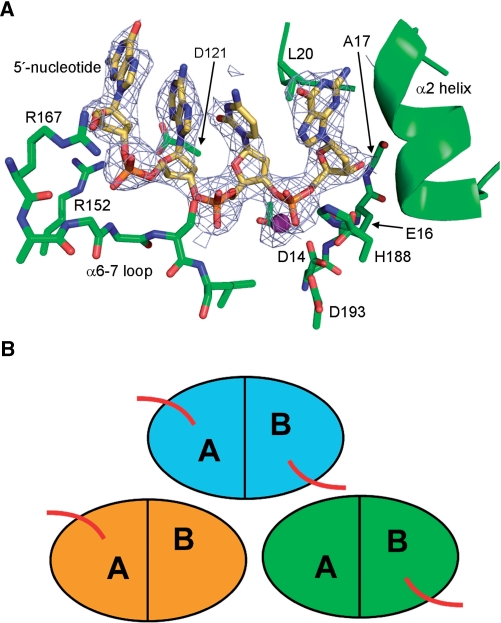
Figure 1.
TREX2 DNA binding. The crystals of the TREX2–DNA complex have three dimers in the asymmetric unit. One of the dimers has ssDNA bound in both active sites while the other two dimers have ssDNA bound in only one of the active sites. (A) Fo-Fc annealed omit difference electron density (1 σ) showing ssDNA density in one of the active sites. Calcium ion shown in magenta. (B) Schematic showing the relative positioning of the three TREX2 dimers in the asymmetric unit. The bound ssDNA is represented by red lines.
The ssDNA is bound in the TREX2 active site through a combination of sequence-independent hydrogen bonding and hydrophobic interactions (Figure 1). A single Ca2+ ion is observed in the active site, coordinated by the conserved acidic residues and scissile phosphate oxygens. The metal ion occupies the canonical position B, leaving position A unoccupied [for a review see (24)]. Similar to the TREX1–DNA complex (10), the terminal nucleotide is anchored in the active site by hydrogen bonds between the 3′-hydroxyl and a carboxylate oxygen of Glu16 as well as the backbone amide nitrogen of Ala17. The nucleotide base is stabilized in a hydrophobic pocket between Leu20 and the hydrophobic face of helix α2. The phosphodiester backbone of the 4 nt are held to the protein through the interaction of Arg152 and hydrogen bonds with the backbone amide nitrogens of the distal end of the α6-α7 loop (amino acids 170–172). Unlike the TREX1–DNA complex in which Arg128 is twisting the base of the 5′-nucleotide into a syn conformation, the equivalent residue, Asp121, in TREX2 is unable to carry out a similar interaction, leaving the nucleotide in the anti conformation.
We have previously shown that the TREX2 exonuclease cooperatively binds DNA and that there is an interdependence of activity in the dimer (15). Catalytic inactivation of one of the monomers (TREX2H188A/TREX2wt) in the dimer showed a 7-fold decrease in exonuclease activity, indicating communication across the dimer interface. It would be expected that if each of the monomers within the dimer operated independently, inactivation of a single monomer would produce only a 2-fold decrease in activity. By comparing our current structure of the TREX2 protein in two DNA binding modes with the existing structure of TREX2 in the apo form (11), we observe conformational rearrangement of residues localized around the active site that help explain the interdependence of activity between the monomers (Figure 2). A superposition of the TREX2 dimer in the unbound form, with either the single DNA bound and double DNA bound form has an RMSD of 0.95 or 0.96 Å, respectively (calculated using Cα atoms). In the absence of bound DNA and divalent ions two specific regions of the protein are disordered—residues 158–169, comprising the arginine-rich DNA binding loop, and residues 185–188 that contain the catalytic residue His188 (11). When the DNA binding loop of one monomer of the TREX2 dimer interacts with DNA to position it in the active site, we see an ordering of residues 165–169 as well as residues 185–188. Further ordering of amino acids 158–164 may happen in the presence of longer oligonucleotide substrates. What is most notable about these structures is that even with a single oligonucleotide binding the dimer ordering of these residues takes place in both monomers. We also observe a 1.8 Å shift of helix α7 towards the active site, better positioning the δ+ dipole moment of the helix to contribute to DNA stabilization (Figure 2). The shift of the α7 helix also moves the amide nitrogen of Tyr170 and Leu172 in the preceding loop to within hydrogen bond distance of the DNA phosphate groups. Previous kinetic studies of TREX2 have demonstrated the catalytic role of His188, and presumably ordering of the loop containing H188 is required for efficient nucleotide hydrolysis (11). Together, the result of these conformational changes appears to provide a significant increase in DNA interactions that likely enhance substrate binding.
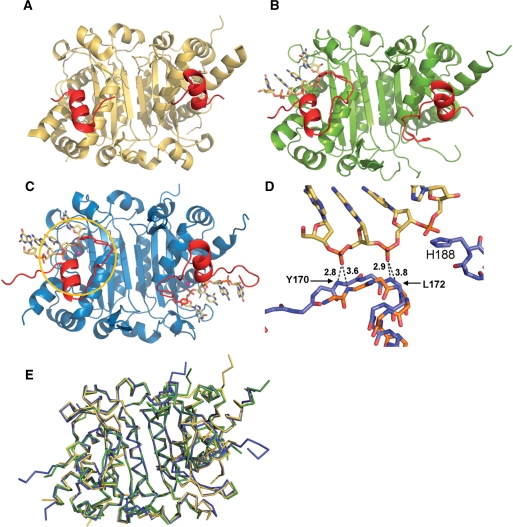
Figure 2.
TREX2 active site conformational change upon DNA binding. A comparison of the TREX2 structures with (A) no DNA bound, (B) one ssDNA oligonucleotide bound to one active site in the dimer and (C) ssDNA bound to both active sites in the dimer. Binding of ssDNA to one monomer induces a conformational change in the active site of both monomers (affected residues shown in red). This includes ordering of DNA binding residues and residues 185–188, which contains the catalytic H188 (shown in gold circle). (D) A superposition of TREX2 bound to DNA (blue) onto the apo TREX2 structure (gold) shows there is also a shift of helix α7 in both monomers that brings the helix closer to the DNA and allows the backbone amide nitrogens of residues 170 and 172 move within hydrogen bonding distance. (E) Superposition of TREX2 dimers in apo (gold), single (green) and double DNA bound (blue) states. The single and double bound states have an RMSD of 0.95 and 0.96 Å, respectively, with the unbound protein. The structures are extremely similar in the dimer interface region with the major structural shifts occurring at the active sites.
Based on these observations, we can propose a model for TREX2–DNA binding and activity in which the monomers function in a cooperative and synergistic fashion with each other (Figure 3). First, when DNA is engaged by the arginine-rich binding loop of either monomer in the dimer a conformational change is induced that results in tighter binding of the 3′-terminus of DNA in the active site of that monomer. This is consistent with the biochemical data that show TREX2 cooperatively binds DNA and the cooperativity exists within each monomer (15). The conformational change due to DNA binding in one monomer is then structurally transmitted to the opposing monomer which also adopts the same conformation and may now also have a higher affinity for substrate. In other words, binding of a DNA 3′-terminus in the active site by one monomer increases substrate affinity of both the monomers. If indeed the ordering of an opposing active site allows increased 3′-terminus binding and higher catalytic efficiency, this model would then imply that DNA binding in the active site is the rate limiting step in catalysis, which is also consistent with previous kinetic measurements (15).
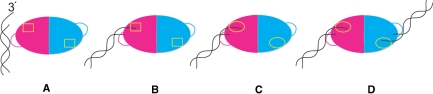
Figure 3.
Model for TREX2 DNA binding. (A) Substrate DNA is recognized by arginine-rich DNA binding loop. (B) The loop positions the 3′-end of the substrate into the active site (yellow rectangle). (C) Presence of ssDNA in the active site induces a conformational change in the active site (yellow oval) of both monomers. (D) Both monomers now have a higher affinity for DNA and increased catalytic efficiency.
The greater than 2-fold loss in activity observed upon mutation of the catalytic His188 to alanine within the mutant/wild-type heterodimer suggests that this residue participates in the apparent communication of activity between the monomers of the TREX2 dimer. It also suggests that the 185–188 loop plays a role in the increased affinity for the 3′-terminus, and if the loop fails to become ordered in one monomer then the conformational change in the active site is not transmitted to the opposing monomer. The second monomer would then function without the benefit of increased binding or efficiency, and therefore, at less than half of maximal measured capacity. It is possible that ordering of the 185–188 loop is dependent upon both DNA binding and the side chain of the histidine making interactions in the active site. Previous structural studies of the DEDD-h exonucleases show the conserved histidine side chain making interactions with the nucleophilic water molecule coordinated by the DNA and divalent metal ion in the substrate complexes, as well as with a phosphate oxygen in the product complexes (10,25).
The mode of TREX2–DNA binding in our current structure is a contrast to the structure of the TREX1–DNA complex that showed a saturation of the active sites with substrate when crystallized with an equivalent ratio of ssDNA (9,10). There also did not appear to be active site conformational changes within the TREX1 structure upon binding substrate (9,10). These structural differences suggest differences in mechanism between the two proteins. If the TREX1 active site does not undergo conformational changes upon substrate binding as TREX2 appears to do there would be less of an entropic barrier for hydrolysis. This is consistent with the fact that the TREX1 enzyme is about 10-fold more efficient than the TREX2 protein (14). Additionally, the individual monomers of the TREX1 dimer appear to have equal affinity for DNA. In contrast, the conformational change induced in both active sites of the TREX2 dimer by binding of DNA to one of the active sites indicates there may be an increased affinity for DNA in the unbound site. This suggests that the TREX2 protein may prefer to function with both active sites engaged simultaneously.
DNA strand displacement
Although the TREX exonucleases are very efficient at hydrolyzing ssDNA substrates, it is likely that at least some of the biological substrates recognized by these enzymes are nicked dsDNA or dsDNA ends (1,26). The TREX1 protein was recently shown to participate in the SET complex where it functions to degrade DNA nicked by the NM23-H1 endonuclease. The architecture of the TREX active sites appears optimized to interact with ssDNA. It is likely that when the protein interacts with dsDNA, single-strand displacement is required to provide a suitable substrate end of about 4 nt to properly bind in the active site for catalysis. Our current structure of the TREX2–DNA complex has provided us with additional information about the TREX proteins potential interactions with dsDNA. Due to the symmetry within the crystal, the ssDNA molecules interacting with separate TREX2 monomers are positioned adjacent to one another in an anti-parallel orientation to form a pseudo-dsDNA molecule (Figure 4). With the 3′-terminus of the substrate strand anchored in the active site, the 5′-end of the ‘complementary’ strand abuts the β1-2 loop. This loop contains the conserved Pro21 and Leu20 residues that form part of the active site cleft. The positioning of the proline at the interface of the nucleotide strands suggests it may act to unzip and denature the dsDNA and feed the substrate strand into the active site.
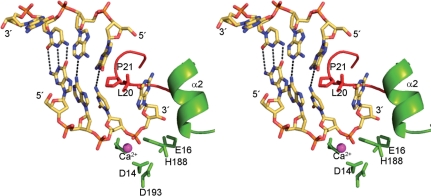
Figure 4.
DNA strand displacement by TREX2. Stereo figure showing how the TREX2 protein potentially interacts with dsDNA. Lattice packing interactions in the crystal created a pseudo-dsDNA that show the 5′-end abutted against the β1-2 loop. This loop which forms part of the substrate binding pocket may play a role in DNA strand displacement to provide ssDNA to the active site from a DNA duplex.
β-Hairpin role in protein–protein interaction
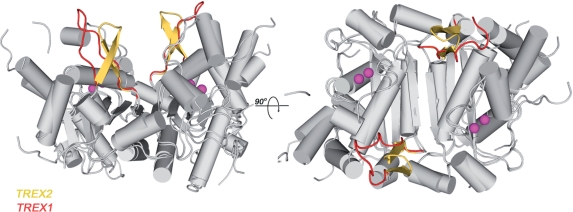
While a biological role for the TREX1 enzyme in the cell has been defined (1,4), the exact function for the TREX2 protein remains unclear. TREX1 is known to participate in the SET complex where it directly interacts with the SET protein and acts to rapidly degrade 3′-ends of nicked DNA during granzyme A-mediated cell death. TREX1 was shown to act within a complex of proteins that include the NM23-H1 nuclease, pp32, HMG-2 and APE1 and to bind directly to the SET protein (1). It is thought that a polyproline II helix (PPH) motif in the TREX1 protein mediates the interactions with the SET complex (9,10). Interestingly, TREX2 was specifically shown not to interact in this complex, again suggesting TREX2 is recruited to a different DNA metabolizing pathway. A superposition of the TREX2 and the TREX1 dimers shows very high similarity overall in the two structures (RMSD = 1.2 Å, Cα atoms using pdbid 1Y97 and 2IOC). The single major difference between the two, however, is at the position of the PPH at the TREX1 dimer interface that is instead a β-hairpin in the TREX2 structure (Figure 5). The PPH forms a distinct three-sided, left-handed helix and is ideally situated to mediate interactions between TREX1 and other proteins allowing positioning adjacent to the exonuclease active site. The β-hairpin in TREX2 is in the identical position as the PPH in TREX1 and protrudes from the surface of the dimer in a similar fashion; suggesting that it may act in a similar capacity to mediate protein–protein interactions. This subtle difference in structure between the two proteins may provide the selectivity switch that allows the recruitment of each exonuclease to the appropriate complex.
Figure 5.
TREX2 and TREX1 protein interactions. A superposition of the TREX1 and TREX2 protein structures shows they are very highly similar. One of major differences in the two structures is that the TREX2 protein contains a β-hairpin (shown in yellow) motif adjacent to the dimer interface, while the TREX1 protein contains a PPH at the equivalent position (red). The PPH is thought to act as a protein–protein interaction motif for TREX1, suggesting that the β-hairpin of TREX2 might serve a similar function in TREX2 but providing differing specificity. Calcium ions from the TREX1 active sites are shown in magenta.
In conclusion, the TREX2–DNA complex has revealed new information about the interaction of this enzyme with substrate. We have observed a conformational change in the enzyme active site upon DNA binding along with a sequential mode of binding that is distinct from that seen with the TREX1 protein. A structural motif in the active site has been identified that may play a role in strand displacement of dsDNA to provide a single-stranded substrate for the enzyme. The unique structural aspects identified in the TREX2–DNA complex compared to the TREX1–DNA complex predict novel biological roles for these two closely related 3′-exonucleases.
FUNDING
American Cancer Society Grant (RSG-04-187-01-GMC to T.H.); National Institutes of Health Grant (RO1 GM069962 to F.W.P.). Funding for open access charge: American Cancer Society (TH).
Conflict of interest statement. None declared.
REFERENCES
- 1.Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino FW, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 3.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O’Hara A, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J. Biol. Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- 6.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′–>5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 7.Perrino FW, Miller H, Ealey KA. Identification of a 3′–>5′-exonuclease that removes cytosine arabinoside monophosphate from 3′ termini of DNA. J. Biol. Chem. 1994;269:16357–16363. [PubMed] [Google Scholar]
- 8.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Brucet M, Querol-Audi J, Serra M, Ramirez-Espain X, Bertlik K, Ruiz L, Lloberas J, Macias MJ, Fita I, Celada A. Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains. J. Biol. Chem. 2007;282:14547–14557. doi: 10.1074/jbc.M700236200. [DOI] [PubMed] [Google Scholar]
- 10.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J. Biol. Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 11.Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3′ –> 5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J. Biol. Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 12.Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 13.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′ –> 5′ exonucleases. Characterization of the recombinant proteins. J. Biol. Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 15.Perrino FW, de Silva U, Harvey S, Pryor EE, Jr., Cole DW, Hollis T. Cooperative DNA binding and communication across the dimer interface in the TREX2 3′-5′ exonuclease. J. Biol. Chem. 2008;283:21441–21452. doi: 10.1074/jbc.M803629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrino FW, Krol A, Harvey S, Zheng SL, Horita DA, Hollis T, Meyers DA, Isaacs WB, Xu J. Sequence variants in the 3′ –> 5′ deoxyribonuclease TREX2: identification in a genetic screen and effects on catalysis by the recombinant proteins. Adv. Enzyme Regul. 2004;44:37–49. doi: 10.1016/j.advenzreg.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55(Pt 10):1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60(Pt 12, Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure (Camb) 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 26.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J. Biol. Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]