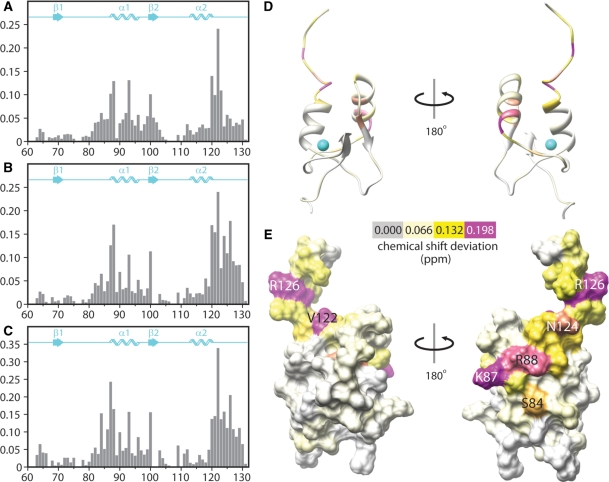
Figure 5.
Chemical shift perturbations of SAP30 ZnF induced by oligodeoxyribonucleotides. Chemical shift deviations induced by (A) a single-stranded 15-mer oligonucleotide, (B) a 10 bp duplex and (C) a 20 bp duplex graphed for each residue in the SAP30 ZnFmotif. Composite deviations of backbone amide proton and nitrogen chemical shifts for each residue were calculated using the formula:  Note that the scale for the y-axis in (C) is different from those in the other panels. Chemical shift deviation data from (C) mapped onto (D) a ribbon representation and (E) the molecular surface of the SAP30 ZnFmotif. These views share the same orientation as those shown in Figure 3. The magnitude of the deviations is color coded according to the key provided; colors are linearly interpolated for intermediate values. Some of the most perturbed residues are annotated.
Note that the scale for the y-axis in (C) is different from those in the other panels. Chemical shift deviation data from (C) mapped onto (D) a ribbon representation and (E) the molecular surface of the SAP30 ZnFmotif. These views share the same orientation as those shown in Figure 3. The magnitude of the deviations is color coded according to the key provided; colors are linearly interpolated for intermediate values. Some of the most perturbed residues are annotated.