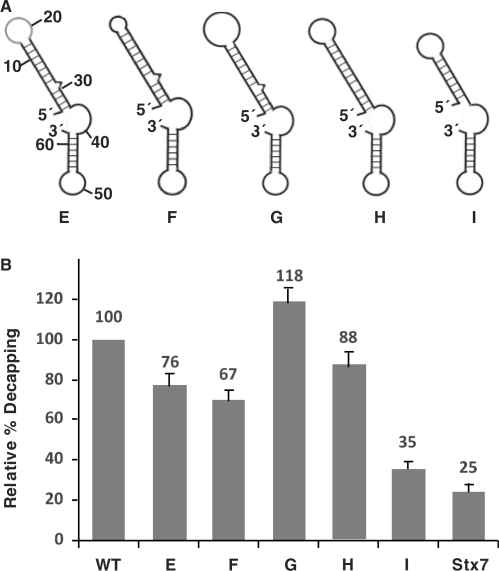
Figure 4.
Mutational analysis of the stem-loop structure. (A) A schematic representation of the predicted structures for the various mutant 2xDE RNAs are shown. WT, wild type; E, substitution of the loop region; F, deletion of the loop to four nucleotides; G, increase the loop to 12 nucleotides; H, deletion of the bulge at nucleotide 29; I, deletion of the lower 4 bp of the stem. (B) Decapping assays were carried out with the indicated mutant 2xDE RNAs and the Stx7 negative control. Decapping efficiencies from three independent experiments were quantitated for each RNA and graphically presented. The decapping efficiency of the wild-type 2xDE RNA was arbitrarily set to 100.