Figure 7.
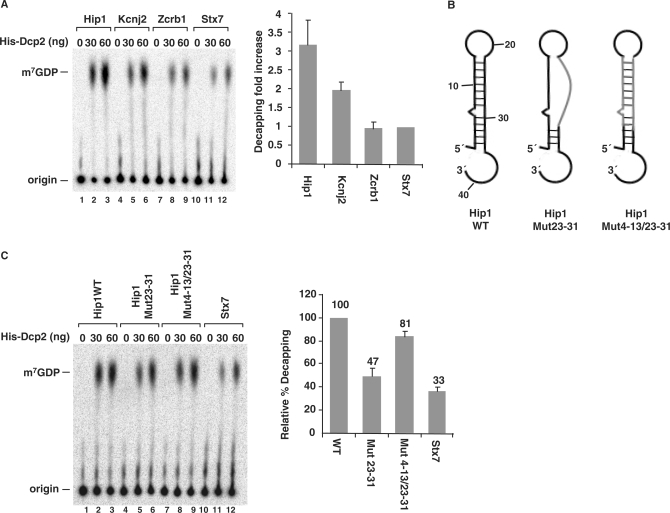
Enhancement of decapping by DBDE-like 5′ end stem-loop structure in cellular mRNAs. (A) Decapping assays were carried out with Hip1, Kcnj2 and Zcrb1 RNAs, which are all predicted to contain potential stem-loop structures at their 5′ end and the Stx7 RNA. Decapping efficiency of Stx7 RNA was arbitrarily set to one and the fold increase relative to Stx7 decapping is plotted. Quantitations were obtained from three independent experiments and the standard deviations are denoted by the error bars. (B) Schematic of the Hip1 5′ UTR wild-type and substitution mutations are shown. WT, wild type; Mut23–31, mutations in nucleotides 23–31 to disrupt the second strand of the stem; Mut4–13/23–31, complementary mutations in both strands of the stem that maintain the same structure. (C) Decapping assays were carried out with the RNAs depicted in B and resolved by PEI-TLC. Labeling is as in the legend to Figure 1A. Quantification of the decapping efficiency for each RNA obtained from three independent experiments are shown on the right with the decapping efficiency of wild-type Hip1 RNA arbitrarily set to 100.