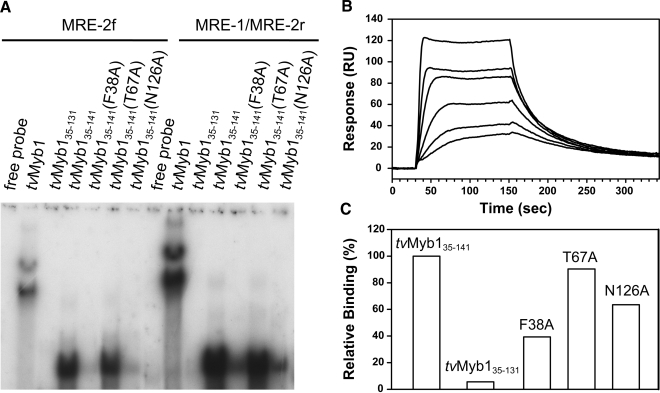
Figure 1.
DNA-binding activity of tvMyb1, tvMyb135–131, tvMyb135–141 and the mutants probed by EMSA (A) and surface plasmon resonance (B and C). In (A), six protein samples, 100 ng tvMyb1 (lanes 2 and 9), 60 ng tvMyb135–131 (lanes 3 and 10), tvMyb135–141 (lanes 4 and 11), F38A (lanes 5 and 12), T67A (lanes 6 and 13) and N126A (lanes 7 and 14) were incubated with -32P-labeled MRE-2f probe (lanes 1–7) or MRE-1/MRE-2r probe (lanes 8–14). All protein/DNA mixtures were separated by 10% polyacrylamide gels by electrophoresis. Free probes were loaded at lanes 1 and 8 as controls. (B) The SPR sensorgrams for the binding of tvMyb135–141 to the DNA duplex. The 16-bp MRE-1/MRE-2r DNA duplex was immobilized on the streptavidin SA sensor chip. The protein concentrations are 25, 12.5, 6.3, 3.1, 1.7 and 0.8 nM. The traces were analyzed with a 1:1 Langmuir-binding model and give an equilibrium dissociation constant (KD) for the tvMyb135–141/DNA interaction to be 1.24 × 10–9 M. (C) Relative DNA-binding ability of tvMyb135–131 and three mutants as calculated from SPR responses 110 s after injection of protein (100 nM) onto the DNA immobilized SA sensor chip.