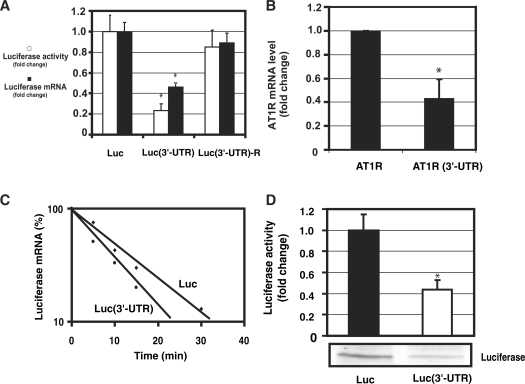
Figure 1.
3′-UTR of AT1R is a negative regulator of AT1R expression. (A) The effect of AT1R 3′-UTR on luciferase activity (white bars). Three different constructs were studied: luciferase, luciferase with AT1R 3′-UTR and luciferase fused with AT1R 3′-UTR in reverse orientation. HEK293 cells were transfected with a luciferase and renilla luciferase plasmids. Cells were lysed and assayed for firefly and renilla luciferase activities. To evaluate mRNA levels, luciferase and GAPDH mRNA levels were measured by quantitative PCR. Corrected luciferase mRNA expression values were calculated by dividing luciferase mRNA by GAPDH mRNA expression (black bars). Values were normalized to the activity or mRNA levels of a luciferase reporter lacking the AT1R 3′-UTR to account for the effect of 3′-UTR. *P < 0.05 versus luciferase without 3′-UTR. (B) The effect of 3′-UTR on AT1R mRNA was evaluated. We compared the mRNA levels of AT1R coding region with or without 3'-UTR. AT1R and GAPDH mRNA levels were determined by quantitative PCR using AT1R coding region specific oligos. AT1R expression was corrected by GAPDH expression. Data were normalized to the activity of AT1R construct lacking the 3′-UTR. The results represent the means ± SD of an average of three independent experiments performed in triplicate for each construct. *P < 0.05 versus AT1R without 3′-UTR. (C) Measurement of the rate of degradation of luciferase mRNA. HEK293 cells were transiently transfected with luciferase construct with or without the full length AT1R 3′-UTR. Transfected cells were then treated with actinomycin D for 5 min, 10 min, 15 min and 30 min. Quantitative PCR was performed with luciferase and GAPDH specific oligos and luciferase mRNA expression was normalized by GAPDH. The mRNA measurements for each construct were normalized to the expression of the construct at time zero. Results are shown as a linear fit. Results represent the means of an average of three independent experiments performed as a triplicate for each construct. (D). Measurement of the rate of luciferase translation in rabbit reticulocyte lysates. In vitro translation of chimeric luciferase constructs with and without 3′-UTR were compared. From the in vitro translation reaction mixture luciferase activity was measured (upper panel) and western blot analysis was performed (lower panel). Translated proteins were detected with streptavidin-HRP detection system. *P < 0.05 versus luciferase without 3′-UTR.