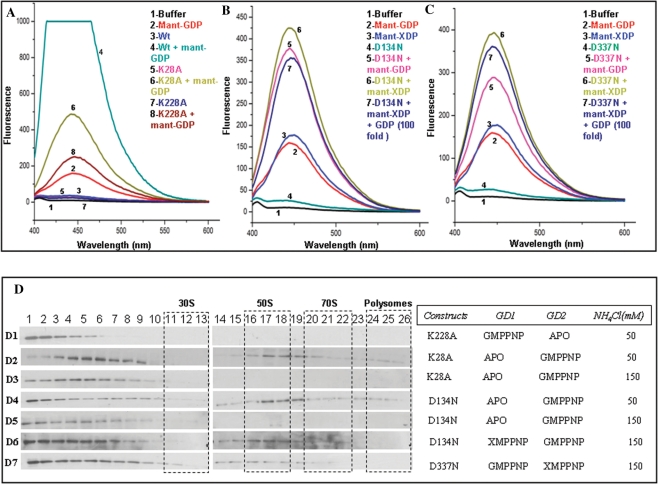
Figure 2.
The importance of GD1 and GD2 in binding 50S. Nucleotide binding was assayed by recording emission spectra (λex 355 nm) (A–C) upon incubating 8 µM of protein with 0.4 µM fluorescent mant-nucleotides (mant-GDP and/or mant-XDP). Spectra corresponding to various nucleotide-bound forms of the protein are indicated in different colors. The protein–nucleotide combinations used are indicated in the insets in corresponding colors and numbers. (A) GDP binding to K28A and K228A mutants is reduced, when compared with the binding to wt-EngA, due to impaired nucleotide binding in the mutated G-domains. (B and C) Proteins carrying D134N and D337N mutations, preferentially bind mant-XDP even in presence of a 100-fold excess of GDP (over mant-XDP). The mutant proteins also bind mant-GDP since the mutation to preferentially bind xanthine nucleotides is created only in one of the G-domains, allowing the other to bind mant-GDP. (D) These mutants were subjected to ribosome co-fractionation experiments as in Figure 1D, with varying salt concentrations and nucleotides, as indicated. (D1) K228A mutant in presence of GMPPNP, which results in EngA[GTP:Apo] state, shows no interaction with ribosomal subunits at 50 mM NH4Cl. Whereas EngA[Apo:GTP] state, generated using K28A, shows an association with 50S (D2) at 50 mM NH4Cl which is abolished (D3) at 150 mM NH4Cl. EngA[Apo:GTP] state was also generated using the mutant D134N. An association with 50S seen (D4) at 50 mM NH4Cl, is abolished (D5) at 150 mM NH4Cl. EngA–50S interaction can be restored (D6) if XMPPNP, too, is provided to the D134N mutant to achieve EngA [GTP:GTP] state. (D7) This effect is verified using D337N mutant, which under similar conditions binds the 50S.