Abstract
The ability to rapidly and efficiently generate reliable Cre/loxP conditional transgenic mice would greatly complement global high-throughput gene targeting initiatives aimed at identifying gene function in the mouse. We report here the generation of Cre/loxP conditional ROSA26-targeted ES cells within 3–4 weeks by using Gateway® cloning to build the target vectors. The cDNA of the gene of interest can be expressed either directly by the ROSA26 promoter providing a moderate level of expression or by a CAGG promoter placed in the ROSA26 locus providing higher transgene expression. Utilization of F1 hybrid ES cells with exceptional developmental potential allows the production of germ line transmitting, fully or highly ES cell-derived mice by aggregation of cells with diploid embryos. The presented streamlined procedures accelerate the examination of phenotypical consequences of transgene expression. It also provides a unique tool for comparing the biological activity of polymorphic or splice variants of a gene, or products of different genes functioning in the same or parallel pathways in an overlapping manner.
INTRODUCTION
Mouse embryonic stem (ES) cells (1,2) have revolutionized the genetic approaches taken in addressing the biological function of mammalian genes. Many have been inactivated or Cre recombinase conditionally mutated by gene targeting. Complementing these loss-of-function approaches many gain-of-function transgenic mouse models have previously been developed by random insertion transgenesis in ES cells or after pronuclear microinjection (3–6). The main limitation associated with random insertion transgenic approaches is the unpredictable variegation of transgene expression (7,8) due to the effect of genomic integration site or transgene copy number (9).
Transgene insertion into the ROSA26 locus (10,11) by targeting has emerged as a favoured solution to gain better control on gene expression. This site provides high targeting efficiency and ubiquitous transgene expression at moderate levels (10,11). The drawback of this approach was the increased complexity of target vector building. More recently, however, several in vitro recombinase-based systems have been developed that have greatly decreased the difficulty of vector construction. The Gateway® DNA cloning technology (12,13) has become popular for cloning DNA elements into plasmids with very high efficiency (90–100%). It consists in the transfer of any sequence (i.e. cDNA) flanked by specific lambda phage integrase recognition sites (attL), into any vector harbouring the corresponding heterotypic (attR) sites via in vitro recombination (LR reactions). In addition, the base pairs at the core of the att recombination site determine their recombination specificity, and mutations within these core sequences have created unique recombination specificity allowing multiple elements to be recombined in tandem (hence the name MultiSite Gateway). This system has been applied in powerful and innovative ways to recombine promoter-based elements with ORF, drug-selectable cassettes and/or reporter markers (14,15).
We present here the efficient generation of Cre/loxP conditional ROSA26 targeting vectors utilizing the ease of Gateway technology. These targeting vectors were used to create several allelic series in the ROSA26 locus of F1 hybrid mouse ES cells. The exceptional developmental potential of these lines allowed the production of germline transmitting chimeras in an extremely efficient, rapid and cost-effective manner by simple aggregation with eight-cell stage embryos. The advancements achieved by combining these tools will significantly reduce the time and costs associated with functional analysis in the mouse.
METHODS AND MATERIALS
Gateway-compatible pEntry and pDestination vector construction
Destination vectors
The Gateway®-compatible pROSA26 Destination Vector (pROSA26-DV1 LMBP 6350) was generated by sub-cloning the attR1-ccdB-chloramphenicol resistance gene (CmR)-attR2 fragment (EcoRV) into a blunt-ended XhoI site of the modified pBigT vector (11) downstream of the SA(splice acceptor)-loxP-PGK-neo-3xpA-loxP cassette and upstream of an IRES-eGFP-pA+ cassette [the IRES-eGFP cassette was previously cloned from the pCALL2-IRES-eGFP vector (a gift from Dr. Corrine Lobe) as a XhoI–NotI fragment into the pBigT vector]. This plasmid was subsequently digested with the unique cutters AscI/PacI and cloned into the pROSA26 targeting vector (AscI/PacI) as previously described (10). The MultiSite pROSA26 Destination Vectors (pROSA26-DV3 LMBP 6352 and pROSA26-DV2 LMBP 6351) were made by blunt-end cloning the R4-ccdB-CmR-R3 fragment from a pBigT-R4-ccdB-CmR-R3 plasmid intermediate into the pROSA26 vector that was cut with PacI and blunt-ended. Orientation relative to the 5′ targeting arm was determined by restriction enzyme analysis and DNA sequencing.
pEntry vectors
The attL1-attL2 flanked cDNA pEntry constructs used in combination with the pROSA26 Destination Vectors were generated as described in Supplementary Figure 1 legend. The 5′ pEntry attL4-pCAGG-loxP-βgeo-3xpA-loxP-attR1 vector (LMBP 6354) was constructed by cloning the pCAGG-loxP-βgeo-3xpA+ fragment (ScaI/XhoI) from the pCall2-IRES-eGFP vector into the pEntry L4-R1 vector (BamHI-Blunt ended XhoI). The 3′ pEntry attR2-IRES-eGFP-attL3 vector (LMBP 6353) was constructed by cloning the IRES-eGFP fragment (XhoI/NotI-blunt) from pcCALL2-IRES-eGFP into pEntry R2L3 (SalI/XhoI-blunt). The plasmids that have been assigned an LMBP accession number are available from the BCCM/LMBP Plasmid collection, Department of Biomedical Molecular Biology, Ghent University, Belgium (http://bccm.belspo.be/about/lmbp.php <http://bccm.belspo.be/about/lmbp.htm>; bccm.lmbp@dmbr.ugent.be).
BP and LR reactions
BP and LR reactions were performed using Clonase™ Enzyme Mix (Invitrogen) according to the suppliers’ instructions with some modifications. Briefly for the monosite Gateway LR reactions 100 ng of the pEntry clone and 150 ng pROSA26-DV in the LR Clonase II mix were left ON at RT. For the MultiSite LR reactions, 100 ng of each of the 3 pENTRY vectors were first incubated with the LR Clonase II mix for 7–8 h at RT and 150 ng of the appropriate pROSA26 Destination vector was added with additional Clonase II mix ON at RT.
Bacterial host and transformation
For construction of destination vectors containing the ccdB gene, library efficiency DB3.1™ electro-competent cells (Invitrogen) were used. For the expression clones DH5 alpha™ (Invitrogen) electro-competent cells were used. For bacterial transformation protocols refer to Supplementary Figure 1 legend.
Sequencing and primers
All plasmid sequencing was conducted at the VIB sequencing facility (ABI3730XL, Applied Biosystems). In order to confirm correct sequence of the inserts of pROSA26 derived expression clones the following primers were used. For the monosite pROSA26-DV1-derived expression clones the sequencing primers S1for (ATCATGTCTGGATCCCCATC) S2rev (GGGGCGGAATTCGATATCAAG) were used (see Figure 1A). For monitoring the degree of excision from tissue samples obtained from the non-excised and excised ROSA26-pCAGG-MDM4 mice the G3for (TCGCTACCATTACCAGTTGGT) and G4rev (CTCTGCTAACCATGTTCATGC) and G5for (GTAGGCAGTGTGTGAGTATC) primers were used.
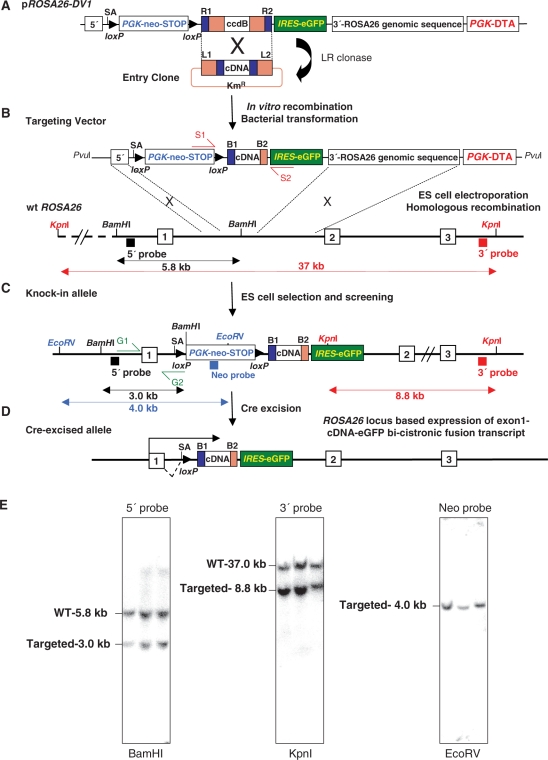
Figure 1.
Generation and analysis of Gateway-compatible conditional ROSA26-promoter-based expression alleles. (A) LR reactions performed between the pROSA26DV-1 vector and cDNA containing pEntry clones for generation of the ROSA26 targeting vector (B). S1 and S2 represent sequencing primers. Black and blue rectangles are the 5′, 3′ external and internal DNA probes. The black double-headed arrows show the expected 5.8-kb (wild-type) and 3.0-kb band sizes (targeted) knock-in (C) alleles in Southern blot analysis using the 5′ external probe on BamH1-digested genomic DNA. Blue arrow shows the expected 4.0-kb band size (targeted) in EcoRV digests using internal neo probe and the red lines depict the 37-kb (wild-type) and 8.8-kb band size (targeted) in KpnI digests using the external 3′ probe. (D) Cre-mediated deletion of the intervening loxP flanked PGK-neo-3xpA (STOP) cassette results in the ROSA26-locus-based expression of an exon1-cDNA-IRES-eGFP bi-cistronic fusion transcript (SA-splice acceptor). (E) Example of Southern blot confirmation of 3 PCR positive ROSA26-Snai2 based ES cell clones used to confirm 5′ integration (left), 3′ integration (middle) and single-copy integration (right).
PCR and Southern analysis
PCR-based screening of targeted ROSA26 ES cell clones was performed using the external G1for primer (TAGGTAGGGGATCGGGACTCT) and the internal G2rev primer (GCGAAGAGTTTGTCCTCAACC) to generate a 1.3-kb PCR fragment (Supplementary Figure 1A). PCR positive clones were confirmed by Southern blotting for 5′ integration using the 5′ external probe (550 bp) and BamH1 digests of genomic DNA (5.8-kb wt allele and 3.0 kb targeted allele). 3′ integration was confirmed using the 3′ external probe (800 bp) and Kpn1 digests (37-kb wt allele and 8.8-kb targeted allele). Both the 5′ and 3′ external Southern probes were generated and provided as a generous gift by Michael Taschner and Dr. Christine Hartmann (IMP, Vienna). An internal Neo probe was used with EcoRV digests to detect a single 4.0-kb ROSA26-targeted allele. For the MultiSite pCAGG promoter targeting experiments to the ROSA26 locus in the sense/anti-sense orientation double EcoRI/KpnI digests were performed and the 5′ external probe generated fragment lengths of 5 kb and 6 kb for targeted events for the anti-sense and sense orientation respectively and an 11-kb fragment for the non-targeted wild-type allele. Similarly, the 3′ external probe generated an 8.8-kb and 9-kb fragment for the anti-sense and sense orientation respectively for the targeted allele and an 11-kb fragment for the wild-type allele. An internal eGFP probe was used to generate a single 5-kb and 6-kb fragment for the anti-sense and sense orientation respectively indicative of single-copy integration.
ES cell culture and aggregations
ES cell culture
The G4 ES cell line was grown and manipulated as previously described (16). Briefly, the G4 ES cells were always grown and manipulated at 37°C in 5% CO2 on mitomycin C-treated mouse embryonic fibroblasts [derived from TgN (DR4)1 Jae embryos] in high-glucose DMEM (Invitrogen), supplemented with 15% ES cell-grade FBS (HyClone), 0.1 mM 2-mercaptophenol, 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids and 2000 U/ml recombinant LIF (DMBR/VIB Protein Service facility). Further expansion and manipulation of these ES cell clones is summarized in Supplementary Figure 1 legend. Parental ES cells were electroporated with the pCAGG-NLS-Cre, PGK-puro plasmid (gift from Dr. Corinne Lobe) and selected according to the protocol outlined in Supplementary Figure 1 legend.
Aggregations
Chimaeras were generated by aggregation of ROSA26-targeted ES cells with CD1 and/or Swiss eight-cell stage embryos as described previously (17). The entire procedure is summarized in Supplementary Figure 1 legend.
RNA extraction and quantitative (q)RT-PCR
Total RNA was extracted from ES cells or polytron-homogenized tissues with TRIZOL (Invitrogen) and purified using phenol/chloroform or RNeasy mini columns (Qiagen). DNaseI treatment was performed at 37°C for 30 min, followed by cDNA synthesis (Superscript II RNase H reverse transcriptase, Invitrogen) starting from equal amounts of RNA. Q-RT-PCR was performed on a LightCycler® 480 system (Roche) using SYBR Green I or Probe Master kit (Roche). Gene expression was normalized for at least two of the following reference genes: hypoxanthine-guanine phosphoribosyltransferase (HPRT), b-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hydroxymethylbilane synthase (HMBS), and results were considered valid only when consistent with multiple internal controls. The primers and probes used for Snai1, Snai2, MDM4, eGFP were as follows: Snai1for: CAGGACTCTAATCCAGAGTTTACCTTC, rev: GGGATGGCTGCCAGCA, probe: AGCAGCCCTACGACCAGGCCCA; Snai2for: GCCAAACTACAGCGAACTGGA, rev: TGTGGTATGACAGGCATGGAG; probe: CACATACAGTGATTATTTCCCCGTATCTCTA GFPFor: TGTCGCCCTCGAACTTCAC; rev: GAGCGCACCATCTTCTTCAAG; GFP-probe ACGACGGCAACTAC; MYC-Mdm4for: AGAAGTTGATCTCCGAGGAGGAT, MYC-Mdm4rev: GCTGTCAGATGCGGAACACT, MYC-Mdm4-probe: CCATTCCACCTCCGCCC.
β-Galactosidase staining, immunohistochemistry and western analysis
The X-gal staining of ES cells and embryos was performed ON at RT as described previously (18). For the E14.5 day embryos staining, they were fist cut sagitally with a razor blade to allow penetration of the X-gal staining reagents. For small pieces of adult tissues, the sections were first whole-mount X-gal stained ON and post-fixed in 4% paraformaldehyde/1X PBS, and paraffin-embedded and sectioned (6–7 µm). Sections were counterstained with Eosin. For immunohistochemical analysis E14.5 embryos were fixed overnight in 4% paraformaldehyde/1X PBS, paraffin-embedded and sectioned (6–7 μm). Slides were stained with antibodies against c-myc tag (1/300, 9E10; Abcam) and GFP (1/300, rabbit IgG fraction; Invitrogen). Detection was performed with the Envision Kit (DAKO) according to the manufacturer's instructions. Sections were counterstained with hematoxylin. For western blot analysis ES cells, embryonic and adult tissue samples were dissolved in standard lysis buffer (pH 8.0) containing phosphatase and protease inhibitors (Complete; Roche Diagnostics), sonicated for 10 s and centrifuged. Protein concentration was measured with the BioRad DC kit. Forty micrograms of each sample was loaded on 8–10% SDS–polyacrylamide gels. Proteins were transferred onto Immobilon-P membranes (Millipore) and blocked with 5% non-fat dry milk, 0.1% Tween-20 in Tris-buffered saline buffer (pH 7.4) prior to incubation with primary antibody. Antibodies were obtained from the following companies: β-galactosidase (Molecular Probes, A-11132), E-cadherin (R&D Systems, MAB 1002), myc (9E10, Abcam), Mdm4 (Sigma, M0445) and Vinculin (Sigma V9131). Detection was done using horseradish peroxidase-coupled secondary antibodies (Sigma; 1:10000) and Western Lightning chemiluminescence reagent (Perkin Elmer).
RESULTS
Conditional ROSA26 targeting vectors compatible with Gateway site-specific recombination cloning
To streamline the production of conditional ROSA26 targeting vectors, we have generated a Gateway ROSA26 destination vector (pROSA26-DV1) as a substrate for the efficient insertion of cDNAs available as attL1-cDNA-attL2 entry clones. The Gateway site in the destination vector is located 3′ of a splice acceptor (SA) followed by a loxP flanked (floxed) PGK-neo-3xpA (STOP) cassette (11) and 5′ of an IRES-EGFP reporter. LR reactions yield pROSA26-DV1-derived expression vectors (top line, Figure. 1). Correct ROSA26 locus targeting was initially screened by Southern blot analysis of genomic DNA using a 200 bp 5′ external probe, previously described for screening ROSA26 recombination events (10). In our hands, this probe gave variable results in Southern blots performed on 96-well plate based genomic DNA restriction analysis. High-throughput screening to identify targeted ES cells was subsequently used in 96-well plate PCR based screens designed to first identify 5′ targeted clones (primers G1 and G2 in Figure 1b, Supplementary Figure 1). We subsequently used larger 5′ (550 bp) and 3′ (800 bp) external probes together with an internal neomycin probe (600 bp) to confirm 5′ and 3′ integration, as well as single-copy insertion into the ROSA26 locus from genomic DNA isolated from 24-well plate formats (Figure 1E). Mice made from ROSA26-targeted ES cells have been inter-crossed with ubiquitous or tissue/cell-specific Cre mice.
The multimerized polyadenylation sequence (3xpA) efficiently stops transcription initiated by both the PGK and ROSA26 promoters (11), which we confirmed by observing the lack of EGFP expression prior to Cre-mediated excision of floxed PGK-neo-3xpA (STOP) cassette (Figure 2A). Cre-mediated deletion of the STOP cassette enables the transcription of the ROSA26 exon1-cDNA-IRES-eGFP bi-cistronic mRNA (bottom line, Figure 1). The EGFP reporter marks cells or tissues where the cDNA of interest is expressed during development and could facilitate the genotyping of transgenic embryos. Here we show embryos resulting from crosses between mice that carry conditional ROSA26-targeted alleles and three different Cre transgenic lines: Sox2-Cre (19) for early overall excision in embryo proper (Figure 2A), and Nestin-Cre (20) or Col2-Cre (21) for excision restricted to the nervous system and to bone/cartilage respectively (Figure 2B and C).
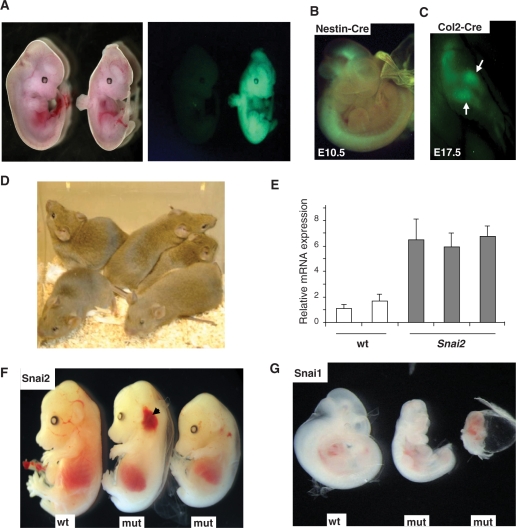
Figure 2.
Generation and analysis of ROSA26-promoter-based mice. (A) Bright field (left panel) and fluorescence (eGFP) images (right panel) of a control non-Cre-excised ROSA26-Snai2-IRES-eGFPTg/+ embryo (left) and mutant Sox2-CreTg/+, ROSA26-Snai2-IRES-eGFPTg/+ embryo (right) showing ubiquitous eGFP expression. (B) Merged fluorescence (eGFP) and bright field image of Nestin-CreTg/+, ROSA26Tg/+ embryo showing eGFP expression in brain and neural tube at E10.5. (C) Fluorescence image of E17.5 forelimb of a Col2-CreTg/+, ROSA26-Tg/+ embryos showing eGFP expression in forelimb elements. (D) Image of totally agouti, seemingly 100% G4 ES-cell-derived G0 male mice. (E) Q-RT-PCR analysis performed on two E11.5 wild-type and 3 Sox2-CreTg/+, ROSA26-Snai2-IRES-eGFPTg/+ mutant littermates showing 2-4-fold upregulated Snai2 mRNA expression levels. (F) Wild-type (wt) E14.5 embryo (left) and two mutant (mut) Sox2-CreTg/+, ROSA26-Snai2-IRES-eGFPTg/+ littermates (right) ubiquitously expressing Snai2 showing pallor and cephalic haemorrhage (arrow). (G) Wild-type (wt) control E10.5 embryo compared with two Sox2-CreTg/+, ROSA26-Snai1-IRES-eGFPTg/+ mutant (mut) littermates ubiquitously expressing Snai1 showing severe developmental defects (left panel).
Through recombination of the pROSA26-DV1 destination vector with cDNA entry clones, conditional ROSA26 targeting vectors can be created for ES cell electroporation within 7 to 10 days. To date, we have produced over 20 such vectors carrying different classes of genes coding for growth factors, kinases, transcription factors, extra-cellular matrix-binding proteins and co-receptors (Supplementary Table 1). In our hands, the average LR reaction efficiency ranges from 70 to 100% expected expression vectors recovered in transformed Escherichia coli colonies (data not shown).
On average, the targeting efficiency of the conditional ROSA26 transgenes in the G4 F1 hybrid ES cells (16) was around 23% (Supplementary Table 1). In our optimized pipeline, a cDNA can be transferred from a Gateway pEntry plasmid to fully characterized single-copy ROSA26-targeted ES cell clones within 3 to 4 weeks (see Supplementary Figure 1). Stable conditional transgenic mice can then be generated from these ES cells with a very high efficiency.
In vivo functional analysis of Gateway-compatible ROSA26 alleles using G4 F1 hybrid ES cells in diploid embryo aggregation
We have previously shown that G4 F1 hybrid ES cells yield mice exclusively derived from ES cells with extreme efficiency via tetraploid embryo complementation assays (16). We have now extended this approach and demonstrated that aggregation of Swiss or CD1 morula-stage diploid embryos with our transgenic G4 ES cells can generate live offspring in which 60% of the neonates are at least 30% chimeric as judged by the agouti colour of their coat, which is a trait of ES cell genetic background. Furthermore, and most interestingly, ∼35% of the pups born were completely agouti implying that these mice were largely (if not totally) derived from ES cells (Figure2D and Supplementary Table 1). Furthermore, all completely agouti males from the 12 ROSA26-targeted ES cell clones characterized, transmitted the targeted allele with the expected 50% Mendelian inheritance. Even the lower 30% chimeric offspring all underwent germline transmission in the first litter. Although no formal GPI isozyme (22) analysis has been performed to confirm the coat-colour estimation of ES cell contribution of the 100% agouti colour mice, it is clear that the G4 hybrid ES cells used in this analysis can out-compete the diploid host CD1/Swiss embryos in giving rise to adult tissues and most importantly to the germline of these founder males.
The initial analysis using our transgenic system focused on two Snai gene family members (Snai1 or Snail, and Snai2 or Slug). Snai genes code for transcriptional repressors involved in the epithelial mesenchymal transition (EMT) that takes place during tissue morphogenesis and have been implicated in cancer progression (23,24). Quantitative RT-PCR analysis of Cre-excised ES cell clones showed 2–4-fold increases in Snai2 and Snai1 mRNA compared to endogenous levels (data not shown, Figure 4E). Such increases were also observed in Sox2-CreTg/+, ROSA26Tg/+ double transgenic embryos expressing Snai2 ubiquitously confirmed by fluorescence microscopy for eGFP (right panel, Figure 2A and E). Ubiquitous expression of Snai2 at these modest levels resulted in variable embryonic lethality, with most embryos showing paleness and cephalic haemorrhaging around embryonic day (E) 13.5–15.5 (Figure 2F). On the other hand, similar levels of ubiquitous Snai1 expression resulted in 100% lethality around E10.5 due to severe global developmental malformations (Figure 2G). We are presently analysing the causes of embryonic lethality caused by ubiquitous Snai1/2 expression. Similar moderate expression has also been documented with several other genes including various VEGF isoforms, as well as Mdm4 Cre-excised ROSA26-locus-targeted ES cells (Figure 5a, data not shown).
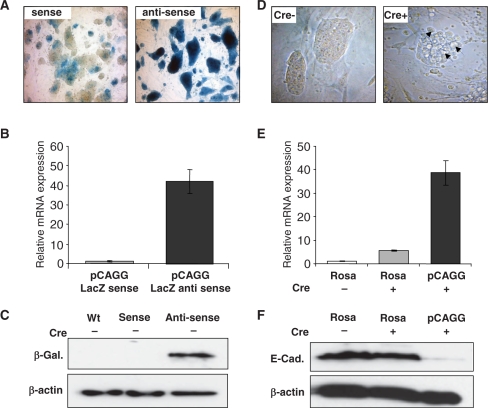
Figure 4.
Analysis of parental and Cre-excised ROSA26-pCAGG-Snai1 ES cell clones. (A) X-gal staining of ES cell colonies of sense (upper-left panel) and anti-sense targeted pCAAG βgeo expressing cells (upper-right panel) (100×). (B) Q-RT-PCR and (C) western blot analysis showing higher β-galactosidase mRNA and protein levels in the anti-sense clones compared to sense clones. β-Actin loading controls (lowest panel). (D) Altered ES cell colony morphology of pCAGG-Snai1 (Cre+) expressing ES cell clones (arrows, top right panel) compared with normal ES cell (Cre–) colony morphology of parental clones (top left panel) (200×). (E) Q-RT-PCR analysis showing elevated levels of Snai1 mRNA transcripts in Cre-excised (Cre+) pCAGG-Snai1 cells compared with ROSA26-promoter-driven Snai1 mRNA levels. (F) Western blot analysis showing decreased E-Cadherin protein levels in ROSA26-Snai1 and pCAGG-Snai1 Cre-excised (Cre+) ES cells, respectively compared to parental ROSA-Snai1 non-Cre excised (Cre–) E-Cadherin levels (middle panel). β-Actin loading controls (lowest panel).
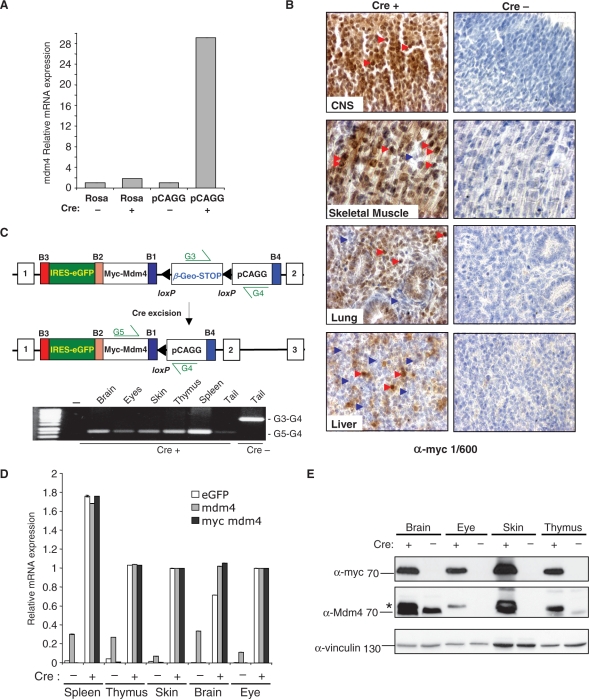
Figure 5.
Analysis of parental and Cre-excised ROSA- and pCAGG-promoter-driven Mdm4 ES cells and pCAGG MDM4 mice. (A) Q-RT-PCR analysis of Mdm4 mRNA expression levels in parental non-Cre-excised (Cre–) clones compared to enhanced Mdm4 mRNA levels of Cre-excised (Cre+) ROSA26 and pCAGG-promoter-driven samples. (B) Immunohistochemical analysis of Sox2-Cre (Cre+) ROSA26-pCAGG-myc-Mdm4 E14.5 tissues using an anti-myc antibody (1/600 dilution) showing widespread cytoplasmic and nuclear localized myc-tagged Mdm4 protein expression in the central nervous system (CNS) and skeletal muscle (red arrow heads) but more mosaic expression in the lung and liver (blue arrow heads) while Cre negative (Cre–) ROSA26-pCAGG-myc-Mdm4 E14.5 tissues show no expression of the myc-tagged Mdm4 protein (400× magnification). Sections were counter-stained with hematoxylin. (C) Schematic of parental (top panel) and Cre-excised (middle panel) ROSA26-pCAGG-Mdm4 alleles. PCR analysis of genomic DNA isolated from control non-Cre Tg (Cre–) pCAGG-Mdm4Tg/+ tail detecting presence of floxed non-recombined allele (G3-G4 primers) but not the excised allele (G5-G4 primers) compared with samples isolated from Sox2-CreTg/+, pCAGG-Mdm4Tg/+ double transgenic organs (Cre+) showing absence of the floxed allele and only excised allele (D) Q-RT-PCR analysis showing widespread and enhanced expression of Mdm4 (grey), myc-tagged Mdm4 (black) and eGFP (white) mRNA transcripts in Sox2-CreTg/+, pCAGG-Mdm4Tg/+ double transgenic tissues (Cre+) compared with non-excised pCAGG-AS-Mdm4Tg/+ controls (Cre–). The relative values are set to zero to accommodate for the lack of eGFP and myc-tagged Mdm4 mRNA expression in the absence of Cre-mediated deletion of the floxed β-geo STOP cassette. (E) Western blot of tissue lysates from Sox2-CreTg/+, pCAGG-Mdm4Tg/+ double transgenic mice (Cre+) showing widespread expression of the myc-tagged Mdm4 protein (upper panel) and absence in pCAGG-Mdm4Tg/+ controls (Cre–). Using an antibody that recognizes both myc-tagged and endogenous Mdm4 protein, the Cre-excised transgenic samples show upregulated myc-tagged Mdm4 protein (*) in all tissues examined compared to endogenous Mdm4 levels. Vinculin protein loading controls (lowest panel).
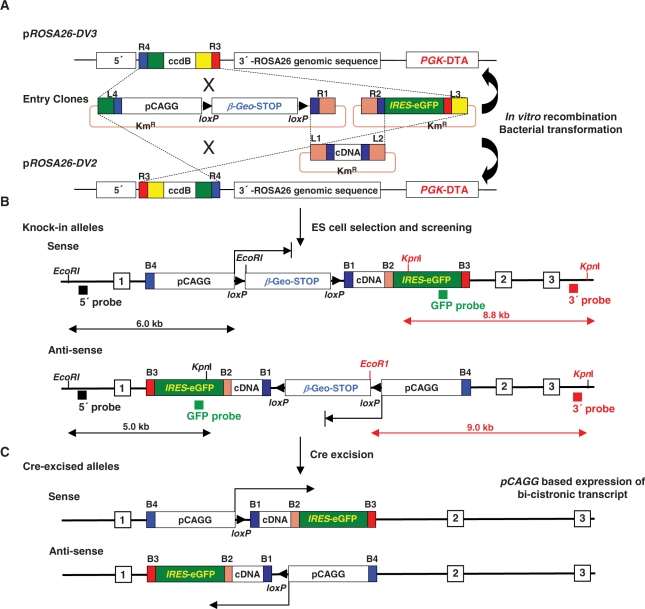
Conditional ROSA26 targeting vectors for higher transgene expression compatible with MultiSite Gateway cloning
ROSA26-promoter-based expression is relatively moderate (see above). In order to obtain higher levels of transgene expression, we designed two MultiSite Gateway destination vectors for the insertion of the strong CAGG promoter (25) floxed βgeo-STOP cassette into the ROSA26 locus, either in sense (pROSA26-DV3) or anti-sense orientations (pROSA26-DV2) (Figure 3). The later configuration was included because the minimal cmv promoter was previously shown to yield higher levels of expression when inserted in the anti-sense versus sense orientation relative to the ROSA26 sense promoter (26). In our MultiSite system, the CAGG promoter floxed βgeo-STOP cassette, the cDNA of interest and the IRES-eGFP reporter cassette are in the 5′, middle and 3′ position, respectively. In both orientations, the βgeo cassette was transcribed before Cre-mediated excision under the control of the CAGG promoter. In cells with the non-excised locus, lacZ in situ staining, β-galactosidase mRNA and protein levels were widespread and significantly higher in the anti-sense configuration (Figure 4a–c).
Figure 3.
Generation of MultiSite Gateway-compatible ROSA26-targeted pCAGG-promoter-based expression alleles. (A) Generation of MultiSite ROSA26 targeting vectors using the 5′-pCAGG-loxP flanked β-geo-3xpA (STOP) cassette, middle-cDNA and 3′-IRES-eGFP reporter pEntry clones. (B) In the conditional knock-in alleles before Cre-mediated excision the pCAGG promoter drives the expression of the β-geo (β-galactosidase-neomycin phosphotransferase fusion gene) cassette (arrows) but not downstream cDNA-IRES-eGFP mRNA expression. Shown are the expected fragment lengths using KpnI/EcoRI digests and the 5′ and 3′ as well as internal eGFP probes used to confirm both sense and anti-sense orientation pCAGG-based single-integration transgenes to the ROSA26 locus. (C) Following Cre-mediated excision of the floxed β-geo cassette, the pCAGG promoter drives cDNA-IRES-eGFP transgene expression in either a sense or anti-sense orientation relative to the sense ROSA26 promoter.
Following Cre-mediated excision of the floxed βgeo-STOP cassette, undifferentiated ES cell colonies in which Snai1 expression was under the control of the CAGG promoter already showed morphological changes (Figure 4d), a result that is consistent with the ability of Snai1 to negatively regulate key adhesion molecules such as E-cadherin (27,28). We next compared mRNA expression driven by the CAGG or the ROSA26 promoter following Cre-mediated excision in ES cells carrying the Snai1 cDNA transgene. The pCAGG-promoter-driven expression was 8–10-fold higher than ROSA26-promoter-based expression (Figure 4E). Enhanced Snai1 expression and altered ES cell morphology was indeed associated with decreased E-Cadherin protein levels that is one of the major epithelial targets repressed by the Snai1 protein (29) (Figure 4F) and these cells were morphologically very similar to E-cadherin deficient ES cell cultures (30).
Next, we determined whether the ROSA26 locus containing the CAGG promoter in the anti-sense orientation could also drive widespread and enhanced transgene expression of Mdm4 in ES cells as well as in adult mice. Similar to the Snai1 expression results, Cre-excised CAGG-promoter-based expression of Mdm4 in the anti-sense orientation was around 10 times higher than ROSA26-promoter-driven expression compared with non-excised parental ES cells (Figure 5A).
Completely agouti males derived from ES cells carrying the conditional ROSA26-pCAGG-Mdm4 allele were inter-crossed either with B6 females to establish the conditional ROSA26-pCAGG-Mdm4 line on a pure B6 background or Sox2-Cre females. Offspring from the later inter-crosses were either analysed at E14.5 for expression of the myc-tagged Mdm4 protein in immunohistochemistry (IHC) analysis or were allowed to develop to term. Double transgenic mice were born in Mendelian numbers and showed no overt phenotypes. This result was somewhat expected, given the predominant role of Mdm4 in p53 regulation and the Mendelian birth ratios of p53 null embryos (31).
IHC analysis of the double transgenic E14.5 embryos from Sox-2 CreTg/+, ROSA26-pCAGG-myc-Mdm4Tg/+ inter-crosses revealed widespread expression of the myc-tagged Mdm4 protein, while no myc-tagged Mdm4 protein was detected on control sections from Cre negative, ROSA26-pCAGG-myc-Mdm4Tg/+ embryos (Figure 5B and Supplementary Figure 2). Closer inspection of the various embryonic tissues showed widespread myc-tagged Mdm4 protein expression localized to the cytoplasm and nucleus in many tissues including the central nervous system and skeletal muscle whereas a smaller subset of tissues showed more mosaic expression of the myc-tagged Mdm4 protein including the lung, liver and especially chondrogenic tissues (Figure 5B and data not shown). A similar expression profile was observed for EGFP in IHC analysis (Supplementary Figure 2E and F). X-gal staining of non-Cre excised ROSA26-pCAGG-myc-Mdm4Tg/+ embryos showed more mosaic β-galactosidase expression (Supplementary Figure 3) that did not completely overlap with the myc-Mdm4 and EGFP IHC analysis (Supplementary Figure 2). In order to investigate the ubiquity of Sox2-Cre-mediated deletion of the floxed β-geo-3XpA+ stop cassette we next performed genomic PCR analysis from adult tissues from Sox-2 CreTg/+, ROSA26-pCAGG-myc-Mdm4Tg/+ mice along with non-excised controls. Genomic PCR analysis with specific primer pairs showed that Sox2-Cre expression resulted in the complete deletion of the β-geo-STOP cassette in a wide range of adult tissues (Figure 5C). Q-RT-PCR analysis performed on the same tissue samples showed expression of Mdm4 and eGFP mRNA transcripts (Figure 5D). Furthermore, western blot analysis using anti-myc-tag or Mdm4-specific antibodies showed myc-tagged Mdm4 protein expression in the same adult tissues (Figure 5E and data not shown). Similar to the developmental analysis, X-gal staining of adult tissue sections showed mosaic β-galactosidase expression (Supplementary Figure 4) that did not completely correspond with the post-excision expression results (Figure 5D and E).
DISCUSSION
The goal of this study was to improve gain-of-function transgenic strategies by targeting cDNAs to a well-defined locus and introducing very efficient in vitro cloning in combination with more broadly applicable ES-cell-based technologies. Here, we have used the ROSA26 locus as a model genomic targeting site to introduce various cDNAs and demonstrate that we could obtain with high targeting efficiency single-copy transgenes conditionally expressed from the endogenous ROSA26 promoter.
The ability of the G4 ES-cell-derived founder males to efficiently transmit the conditional ROSA26-based transgene in the first litter is further expected to decrease the waiting time and number of chimeras that have to be analysed for germline transgene transmission. Such mice can be immediately bred to any Cre line of interest to obtain transgenic animals with modest ROSA26-promoter-driven transgenic expression according to the wide spectrum of constitutive and inducible tissue-specific Cre lines currently available (see http://www.mshri.on.ca/nagy for a partial and ever-expanding list).
An additional benefit of using the Gateway recombination system in tandem with our ROSA26 locus targeting vectors is that there is an ever-growing list of Gateway-compatible pEntry clones (32) as well as commercially and publicly available attB-containing cDNA vectors that can be converted into pEntry clones by performing BP reactions with donor vectors containing the corresponding attP sites (as an example see the following web resource: http://www.imagenes-bio.de/products/sets_libraries/image). These vectors and approaches outlined in this manuscript therefore set the stage for the high-throughput systemized creation of conditional gain-of-function ES cell libraries carrying a wide spectrum of ORF subfamilies. To further increase targeting efficiencies that would be needed for the efficient creation of such gain-of-function resources, the Gateway technologies described here should in the future be merged with in vivo recombinase-based technologies such as [FlpE/Frt (33), and ϕC31/attP systems (34)] to enable recombinase-mediated cassette exchange (RMCE) in vivo to the ROSA26 locus (35,36) as well as other loci (37). These types of RMCE-based, ROSA26-based-targeting approaches have already been demonstrated to be extremely efficient in the creation of RNA polymerase III-based gene knockdowns using gene-specific shRNA sequences (35,38).
Using MultiSite Gateway recombination technologies we have inserted the strong CAGG promoter bi-directionally into the ROSA26 locus in both a sense and anti-sense orientation relative to the ROSA26 sense promoter. We have demonstrated that in undifferentiated ES cells, the anti-sense configuration of the pCAGG promoter is optimal for enhanced expression compared to the sense orientation. This result is consistent with previous reports that the minimal cytomegalovirus (CMV) promoter elements also drive higher transgene expression in the anti-sense orientation in undifferentiated ES cells, a result that is thought to be due to ROSA26 sense promoter interference (26). In our initial analysis of the ROSA26-pCAGG-myc-Mdm4 mice we can clearly see widespread Mdm4 and eGFP protein expression during development (at E14.5) and widespread expression in the adult in Cre-excised tissues. However, we have also documented that there is a significant lack of correlation between pCAGG promoter-driven β-galactosidase expression and X-gal staining in the non-excised situation compared with the broader expression of myc-Mdm4 transgene following Cre-mediated excision of the floxed βgeo STOP cassette. In addition, our initial analysis of Mdm4 protein expression demonstrates that even after Cre-mediated excision of the β-geo STOP cassette we are not getting truly ubiquitous protein expression. For our purposes where we want to mimic the tumour situation in which there is clonal expansion of a small subset of transformed cells, mosaic but high levels of Mdm4 expression may be acceptable or even desirable. However, for studies where ubiquitous expression of the transgene is essential in a given tissue, we propose that the ROSA26-promoter-based expression strategies outlined above are superior when used with well-characterized Cre lines.
Previous attempts to use the CAGG promoter in the generation of the Cre-based bi-reporter Z/AP (39) and Z/EG (18) mice have demonstrated similar findings to those which we have documented in this study. Random integration of the pCAGGs-based vectors into undifferentiated ES cells resulted in only around 10% of single-copy random integrants that showed ubiquitous (>90%) X-gal staining in undifferentiated ES cell clones. These results indicate that the CAGGs promoter is quite sensitive to positional integration effects and raise the possibility of promoter interference from the ROSA26 anti-sense-promoter in a tissue-specific manner (40). As well, the lack of correlation between pre-Cre excision-based expression of β-galactosidase and post-Cre-based expression of reporter constructs (18,39) is similar to our observed results where β-galactosidase expression showed more limited and mosaic expression compared with Cre-mediated expression of myc-Mdm4 protein and EGFP. These results may be attributable to elements within the β-geo cassette interfering with pCAGGs-based expression prior to Cre-excision and/or the fact that the bacterial β-galactosidase gene has previously been demonstrated to show mosaic expression particularly in adult tissues (41,42) that may be related to sub-optimal codon usage of the prokaryote sequences (18) or transgene silencing (41,42).
These studies demonstrate that caution should be taken when inserting exogenous promoters into the ROSA26 locus in both sense and anti-sense orientations and it is still not clear if tissue-specific promoter elements can be used to drive ubiquitous and/or cell/tissue-specific expression from this locus on their own during development and in the adult. Recently, several groups have demonstrated the utility of using insulator sequences in transgenic studies (43). Ciavatta et al. (44) have demonstrated that copy number-dependent use of insulator sequences at the widely expressed HPRT locus can dramatically protect chicken-β-actin-promoter-based EGFP expression during development and in adult tissues. It is envisioned that similar strategies should in the future be used for the targeting of the pCAGG or other tissue-specific promoters to the ubiquitously expressed ROSA26 locus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
The Association for International Cancer Research grant (Scotland #06-0570 to G.B. and J.H.). PhD grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen to L.H.). Belgium Science Policy (to M.N. and S.G., Partial) Interuniversity Attraction Pole (grant P6/30). Funding for open access charge: Ghent University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Frank Costantini, Corrine Lobe and Christine Hartmann for providing reagents used in this study. We would also like to thank Dr. Chris Guerin of the DMBR for his help with microscopy analysis.
REFERENCES
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costantini F, Lacy E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature. 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 5.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl Acad. Sci. USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner EF, Stewart TA, Mintz B. The human beta-globin gene and a functional viral thymidine kinase gene in developing mice. Proc. Natl Acad. Sci. USA. 1981;78:5016–5020. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rijkers T, Peetz A, Ruther U. Insertional mutagenesis in transgenic mice. Transgenic Res. 1994;3:203–215. doi: 10.1007/BF02336773. [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu. Rev. Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- 9.Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC, Evans RM. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300:611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 11.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzen F. Gateway recombinatorial cloning: a biological operating system. Expert. Opin. Drug Discov. 2007;2:571–589. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- 14.Karimi M, Depicker A, Hilson P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007;145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rual JF, Hill DE, Vidal M. ORFeome projects: gateway between genomics and omics. Curr. Opin. Chem. Biol. 2004;8:20–25. doi: 10.1016/j.cbpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 16.George SH, Gertsenstein M, Vintersten K, Korets-Smith E, Murphy J, Stevens ME, Haigh JJ, Nagy A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy AaRJ. In: Gene Targeting: A Practical Approach. Joyner AL, editor. Oxford: IRL Press; 2000. pp. 177–206. [Google Scholar]
- 18.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 19.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;119(Suppl. 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 20.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 21.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- 22.Peterson AC, Frair PM, Wong GG. A technique for detection and relative quantitative analysis of glucosephosphate isomerase isozymes from nanogram tissue samples. Biochem. Genet. 1978;16:681–690. doi: 10.1007/BF00484725. [DOI] [PubMed] [Google Scholar]
- 23.De Craene B, van Roy F, Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signal. 2005;17:535–547. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 25.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 26.Strathdee D, Ibbotson H, Grant SG. Expression of transgenes targeted to the Gt(ROSA)26Sor locus is orientation dependent. PLoS ONE. 2006;1:e4. doi: 10.1371/journal.pone.0000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 28.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 29.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 30.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl Acad. Sci. USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey M, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 32.Zavzavadjian JR, Couture S, Park WS, Whalen J, Lyon S, Lee G, Fung E, Mi Q, Liu J, Wall E, et al. The alliance for cellular signaling plasmid collection: a flexible resource for protein localization studies and signaling pathway analysis. Mol. Cell Proteomics. 2007;6:413–424. doi: 10.1074/mcp.M600437-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 34.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seibler J, Kuter-Luks B, Kern H, Streu S, Plum L, Mauer J, Kuhn R, Bruning JC, Schwenk F. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res. 2005;33:e67. doi: 10.1093/nar/gni065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 38.Hitz C, Wurst W, Kuhn R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007;35:e90. doi: 10.1093/nar/gkm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev. Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 40.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paldi A, Deltour L, Jami J. Cis effect of lacZ sequences in transgenic mice. Transgenic Res. 1993;2:325–329. doi: 10.1007/BF01976173. [DOI] [PubMed] [Google Scholar]
- 42.Ramirez A, Milot E, Ponsa I, Marcos-Gutierrez C, Page A, Santos M, Jorcano J, Vidal M. Sequence and chromosomal context effects on variegated expression of keratin 5/lacZ constructs in stratified epithelia of transgenic mice. Genetics. 2001;158:341–350. doi: 10.1093/genetics/158.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 44.Ciavatta D, Kalantry S, Magnuson T, Smithies O. A DNA insulator prevents repression of a targeted X-linked transgene but not its random or imprinted X inactivation. Proc. Natl Acad. Sci. USA. 2006;103:9958–9963. doi: 10.1073/pnas.0603754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.