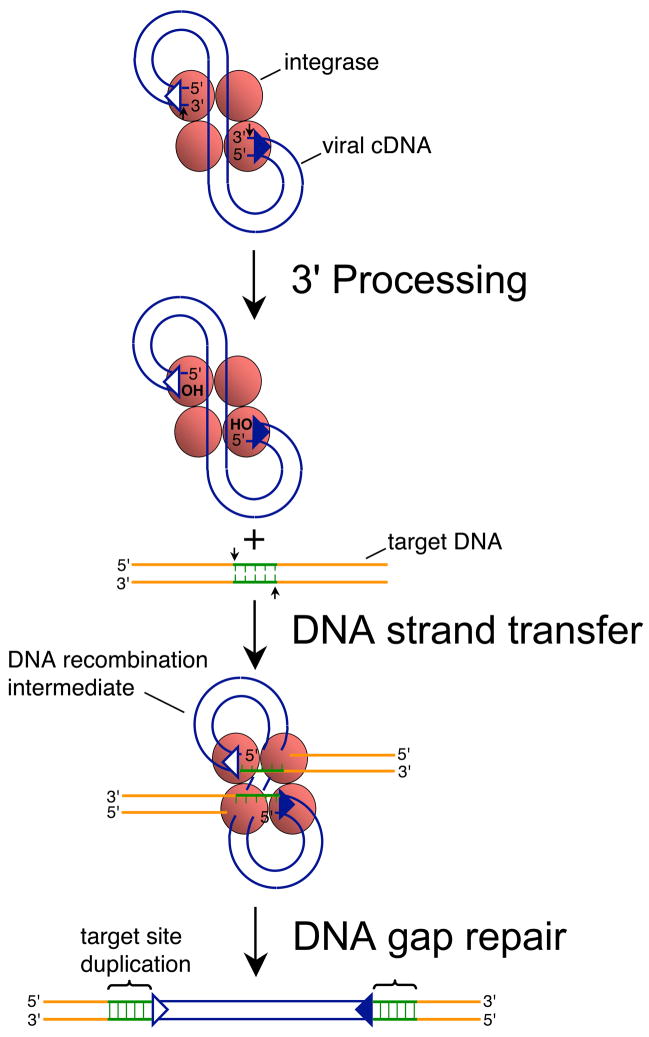
Fig. 1.
Mechanism of retroviral DNA integration. Reverse transcription yields linear cDNA with a copy of U3/R/U5 sequences, also known as the long terminal repeat (LTR), at each end. Integrase recognizes approximately 15–20 bp of each end region [39, 40]. The upstream recognition or attachment (att) site is therefore comprised of U3 sequences (open triangle) whereas the downstream att site is U5 (filled blue triangle). Integrase (red shaded ball) cleaves each att site adjacent to the invariant sequence 5′-CA-3′ during 3′ processing (marked by small vertical arrows), removing the dinucleotide GT from each end of HIV-1 cDNA [4, 6]. Integrase uses the oxygen nucleophiles at the exposed CAOH ends during DNA strand transfer to cut the target DNA at the small vertical arrows, which at the same time joins the viral ends to the resultant 5′-phosphates [6]. The DNA recombination intermediate, with unjoined viral DNA 5′ ends, is the structure formed in in vitro PIC integration assays [1, 2, 5]. DNA repair yields the integrated provirus flanked by the sequence duplication of the target DNA staggered cut (green lines), which is 5 bp for HIV-1 [41, 42]. Integrase functions as a multimer in vitro and in vivo, though the precise make-up of the active multimer during infection is unknown. It is depicted here as a tetramer for simplicity.