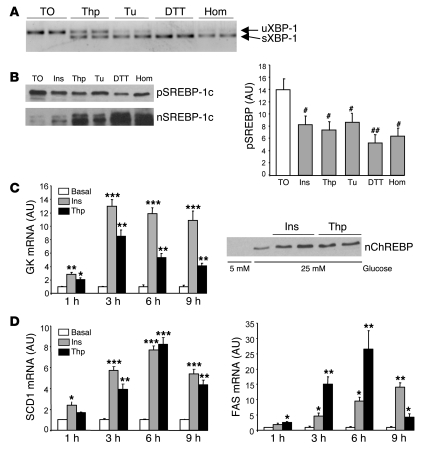
Figure 2. Effect of ER stress on SREBP-1c proteolytic cleavage and SREBP-1c target genes in cultured rat hepatocytes.
(A and B) Hepatocytes were incubated for 6 h in M199 medium supplemented with the LXR agonist TO-901317 (TO; 10 μM) and then treated for 1 h with 100 nM insulin (Ins), 300 nM thapsigargin (Thp), 1 μg/ml tunicamycin (Tu), 500 μM DTT, or 5 mM homocysteine (Hom). (A) Unspliced (u-) and spliced (s-) forms of XBP-1 mRNA, measured by RT-PCR. (B) Immunoblot analysis of SREBP-1c precursor and nuclear SREBP-1c. Quantification of SREBP-1c precursor is shown at right. Results are mean ± SEM of 3 independent cultures of hepatocytes in which 3 plates were collected for each condition. #P < 0.05, ##P < 0.01 versus TO-901317 alone. (C and D) Hepatocytes were treated for 1, 3, 6, and 9 h with 100 nM insulin or 300 nM thapsigargin in medium containing 25 mM glucose. (C) Total RNA from triplicate plates of hepatocytes was analyzed for the expression of GK. For the measurement of nuclear ChREBP, hepatocytes were cultured in the presence of 5 or 25 mM glucose and treated for 3 h in the presence of 100 nM insulin or 300 nM thapsigargin. (D) Total RNA from triplicate plates of hepatocytes was analyzed for expression of SCD1 and FAS mRNA. *P < 0.05, **P < 0.01, ***P < 0.001 versus basal value.