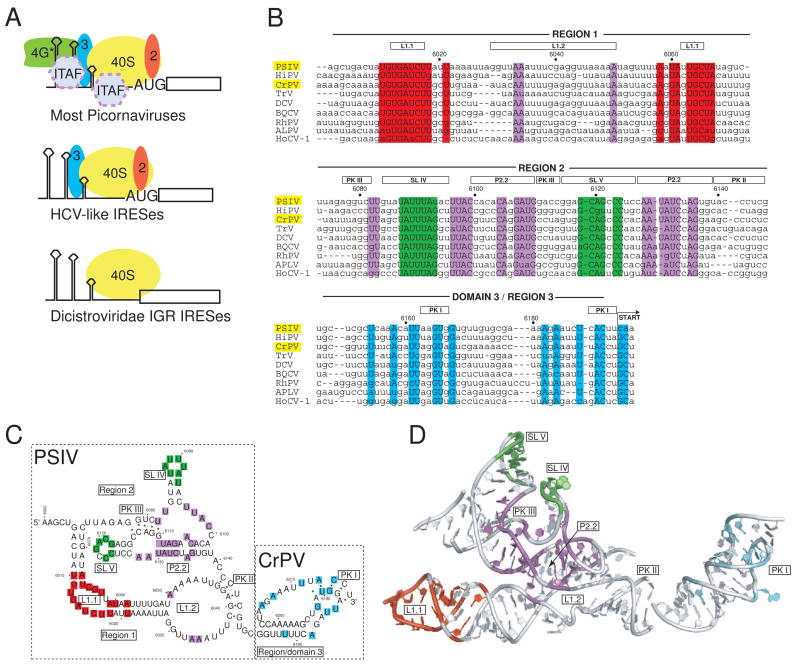
Fig. 1. IRES groups and the IGR IRES RNAs.
a) Cartoon of different strategies used by various IRES groups to recruit a 40S subunit to the IRES RNA. The most complex are those generally associated with the picornaviruses, which require several eIFs and often ITAFS in order to recruit the ribosome. These are subdivided into the entero-/rhinovirus class, the cardio-/apthoviruses class, and the hepatitis A virus class. The HCV-like IRESs (which include some picornaviruses) use a smaller set of eIFs and can bind directly to the ribosome. The most streamlined IRESs are shown at the bottom, which are the IGR IRESs from the Dicistroviridae. These IRESs bind directly to the ribosome, require no eIFs, and do not initiate from a canonical AUG start codon. More atypical IRESs, such as those from the 5′ UTR if the Dicistroviridae, are not shown. b) Sequence alignment of nine members of the Dicistroviridae IGR IRES group, all type 1. The approximate locations of the various regions and key secondary structure elements are indicated above the alignment, and the nucleotide numbers for the PSIV IRES are shown. In this review, I use the term “region” to refer to a logically connected group of secondary structure elements and reserve the use of the term “domain” for a part of the RNA shown to fold into an independent, stably folded three-dimensional structure. Locations in the structure that are highly conserved (sequence is the same in at least 7 of the 9 IRESs shown) are colored. The location of the non-AUG codon that is the start of translation is indicated, and the names of the two IRES RNAs for which crystal structures have been solved are highlighted in yellow. The alignment is based on previously published work (Jan, 2006; Kanamori and Nakashima, 2001). Abbreviations: Plautia stali intestine virus (PSIV), himetobi P virus (HiPV), cricket paralysis virus (CrPV), Triatoma virus (TRV), Drosophila C virus (DCV), black queen-cell virus (BQCV), Rhopalosiphum padi virus (RhPV), aphid lethal paralysis virus (APVL), Homalodisca coagulata virus-1 (HoCV-1). c) Secondary structure model of the ribosome-binding domain (regions 1+2) of the PSIV IGR IRES and the P-site domain (domain 3) of the CrPV IGR IRES, with secondary structure elements that are mentioned in the text labeled. The locations of conserved sequence are colored to match panel 1b. The secondary structure is drawn in a manner that allows it to be more directly compared to the structure shown in panel 1d. d) Combined crystal structures of the ribosome-binding domain of the PSIV IGR IRES (Pfingsten et al., 2006) and the P-site domain of the CrPV IGR IRES (Costantino et al., 2008), with structural elements labeled. Nucleotides that are highly conserved are colored to match panels 1b and 1c. The orientation of the two structures relative to one another matches their location when docked into cryo-EM reconstructions of CrPV IGR IRES-ribosome complexes (Costantino et al., 2008; Schuler et al., 2006; Spahn et al., 2004).