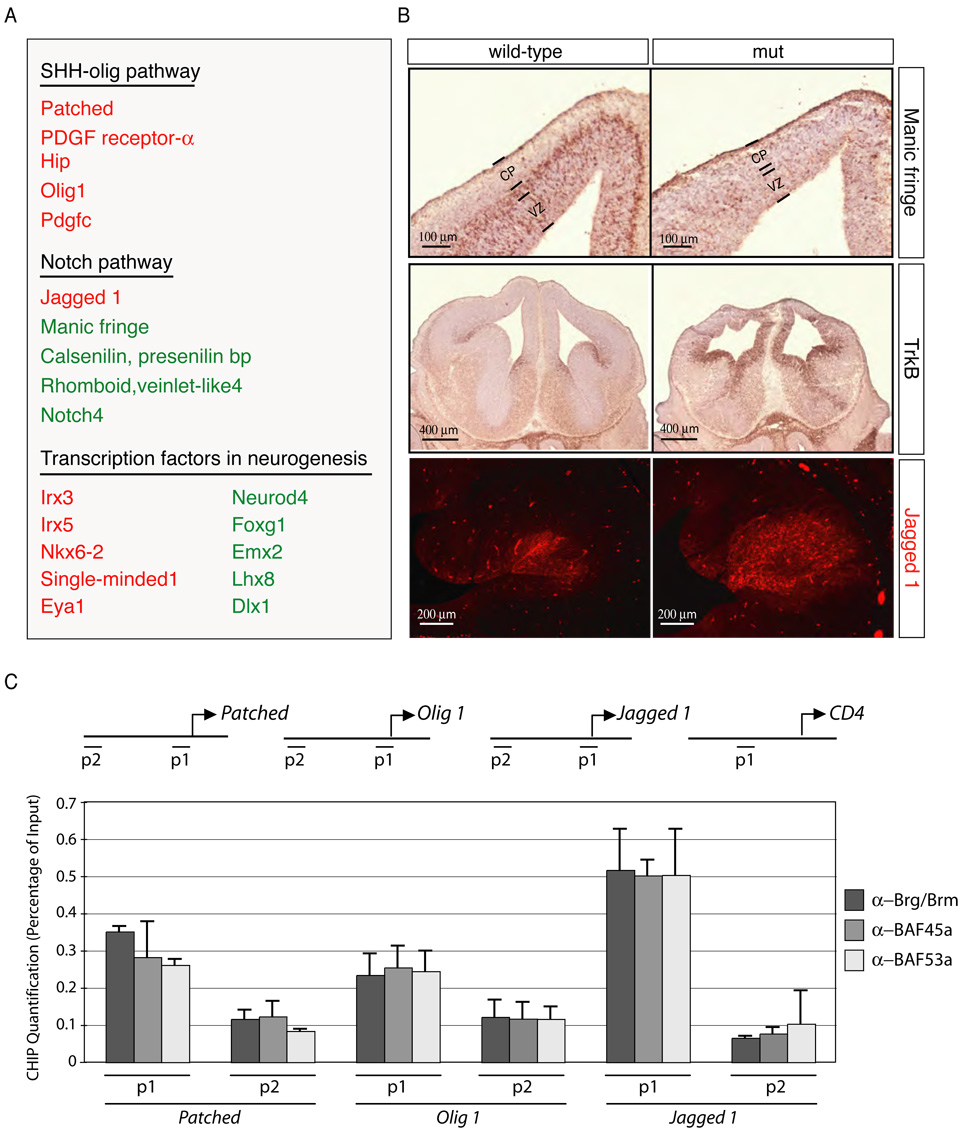
Figure 6. Brg-containing npBAF complexes regulate the transcription of Sonic Hedgehog (SHH) and Notch signaling components in neural development.
A. Microarrays (Mouse 430 2.0 arrays (Affymetrix) containing 39,000 transcripts) were probed with cRNA generated from RNAs isolated from 3 control and 3 Brg-deficient E12.5 telencephalons. Default settings were used to normalize the data and for comparison. Selected genes changed more than two-fold (p<0.01) are shown. Genes that are downregulated in the absence of Brg appear in green and genes that are up-regulated are in red.
B. In situ hybridization and immunostaining analyses of transcriptional targets of npBAF complexes in the E13.5 mouse brain. Positive cells are dark brown (coronal sections). Upper panel: Expression of Manic fringe in newly-born neurons requires Brg. VZ: ventricular zone; CP: cortical plate. Middle panel: Repression of TrkB expression in neural progenitors requires Brg. Lower panel: Increased Jagged1 protein levels in Brg-deficient basal forebrain.
C. BAF45a, BAF53a, Brg and npBAF complexes are present at the promoters of components of the SHH and Notch signaling pathways (P1). P2 regions are ~5 kb upstream. Anti-Brg/Brm, BAF45a and BAF53a antibodies were used for chromatin immunoprecipitation from E13.5 mouse forebrain lysates followed by quantitative PCR. A genomic element of the murine CD4 gene was used as a negative binding control (Chi et al., 2002). The relative binding abundance of each region is presented as percentage of the input DNA material. The data were normalized to the relative abundances of the CD4 P1 region and DNA precipitated by an anti-GFP antibody (negative control) (n=3 independent experiments). Primer sequences are available upon request.