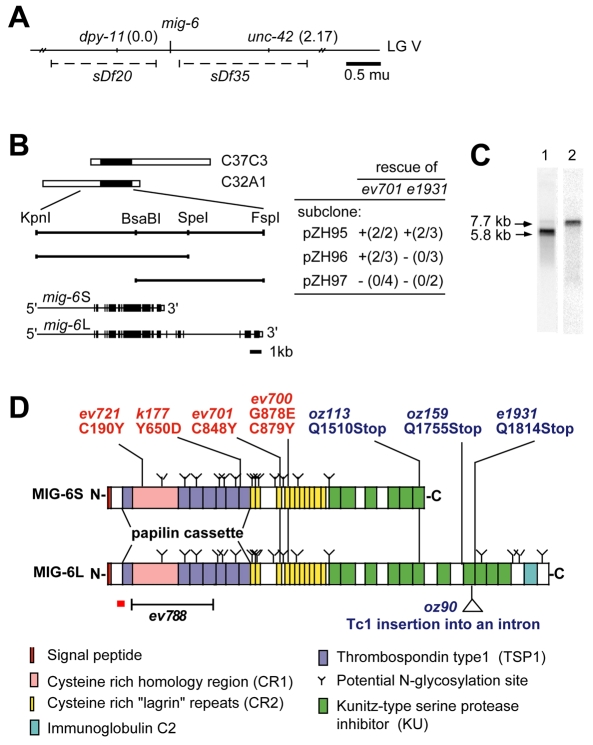
Fig. 3.
Cloning and characterization of the mig-6/papilin gene. (A) mig-6 was mapped to the region between two deficiencies, sDf35 and sDf20, on LG V. (B) Schematic drawing of cosmid clones (white bars with single black region denoting the mig-6 gene) and subclones (unbroken lines representing the KpnI and FspI fragments), with rescue results with these DNA fragments shown on the right and exon/intron organization of mig-6 shown below (exons and 3′ UTRs are black and white boxes, respectively). Rescue of mig-6 class-s (ev701) lethality and mig-6(e1931) sterility were determined as described in the Materials and methods. These experiments were internally controlled by scoring transgenic Dpy animals and their non-transgenic Dpy siblings. The number of rescuing transgenic lines over the number of lines tested is indicated in parentheses. DTC defects, which in mig-6(s) and mig-6(l) alleles manifest as clear patches in the body, were also rescued (denoted `+'). (C) Northern blot analysis of 3 μg of mRNA from a mixed-stage population of wild-type C. elegans. A probe predicted to hybridize to both mig-6S and mig-6L detected two bands (lane 1). Longer exposure with a mig-6L-specific probe (exon 14-17) shows a single band at 7.7 kb (lane 2). (D) Schematic diagrams of predicted protein structures of MIG-6S and MIG-6L with sites of molecular lesions found in class-l (blue) and class-s (red) alleles. The extent of the ev788 deletion is indicated. The region corresponding to the peptide used for raising an antibody is indicated by an underlying red bar.