Abstract
Past population size can be estimated from modern genetic diversity using coalescent theory. Estimates of ancestral human population dynamics in sub-Saharan Africa can tell us about the timing and nature of our first steps towards colonizing the globe. Here, we combine Bayesian coalescent inference with a dataset of 224 complete human mitochondrial DNA (mtDNA) sequences to estimate effective population size through time for each of the four major African mtDNA haplogroups (L0–L3). We find evidence of three distinct demographic histories underlying the four haplogroups. Haplogroups L0 and L1 both show slow, steady exponential growth from 156 to 213 kyr ago. By contrast, haplogroups L2 and L3 show evidence of substantial growth beginning 12–20 and 61–86 kyr ago, respectively. These later expansions may be associated with contemporaneous environmental and/or cultural changes. The timing of the L3 expansion—8–12 kyr prior to the emergence of the first non-African mtDNA lineages—together with high L3 diversity in eastern Africa, strongly supports the proposal that the human exodus from Africa and subsequent colonization of the globe was prefaced by a major expansion within Africa, perhaps driven by some form of cultural innovation.
Keywords: mitochondrial DNA, Africa, phylogenetics, Bayesian skyline plot, coalescent theory
1. Introduction
Phylogenetic analyses of human genetic diversity are revealing an increasingly detailed picture of the human colonization of the globe (Forster & Matsumura 2005; Macaulay et al. 2005; Thangaraj et al. 2005; Olivieri et al. 2006; Witherspoon et al. 2006; Underhill & Kivisild 2007; Atkinson et al. 2008). A Late Pleistocene African origin of modern humans is now widely accepted, and is supported by mitochondrial DNA (mtDNA; Cann et al. 1987; Vigilant et al. 1991; Watson et al. 1997; Ingman et al. 2000), Y-chromosome (Underhill et al. 2000; Underhill & Kivisild 2007) and autosomal (Gonser et al. 2000; Zhivotovsky et al. 2000; Alonso & Armour 2001; Rogers 2001; Marth et al. 2004; Witherspoon et al. 2006) evidence. More recent mtDNA-based work has suggested a human expansion from Africa along the Indian Ocean coast 50–70 kyr ago (Forster & Matsumura 2005; Macaulay et al. 2005; Thangaraj et al. 2005) linked with substantial population growth in South Asia (Atkinson et al. 2008), followed by later expansions into Sahul (Friedlaender et al. 2005; Atkinson et al. 2008), the Levant and North Africa (Olivieri et al. 2006), Europe (Atkinson et al. 2008) and the New World (Forster et al. 1996; Atkinson et al. 2008). Within Africa, however, the nature of population expansions before, during and after the global diaspora remains unclear.
Together with evidence from archaeology and other genetic loci, mtDNA diversity can help elucidate human prehistory in sub-Saharan Africa, just as it has on a global scale. The absence of recombination, combined with a high copy number and fast rates of genetic change, makes mtDNA well suited to phylogenetic analysis over the time scale of modern humans. Inferences must be made with the caveat that, by virtue of being inherited from mother to child, mtDNA can only capture the history of the maternal lineage. In addition, as with any single-locus analysis, parameter estimates are only valid to the extent that the locus reflects population demographic history and not the influence of selection or changes in population structure. However, by combining mtDNA-based findings with evidence from other genetic loci, as well as archaeology and linguistics, we can resolve an increasingly detailed picture of human prehistory.
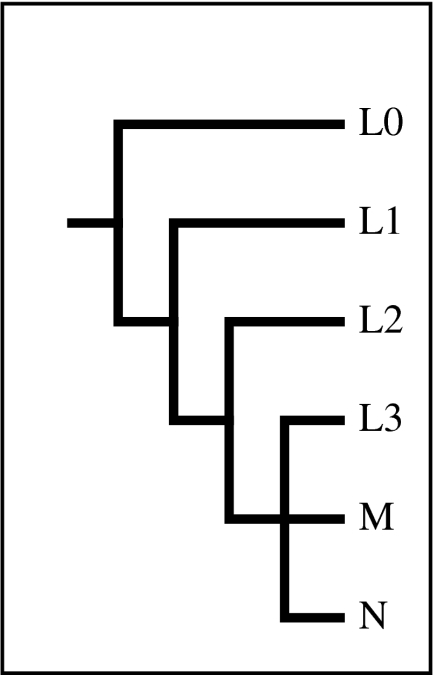
The time to the most recent common ancestor (TMRCA) of the human mtDNA tree is dated to between approximately 150 and 250 kyr ago (Mishmar et al. 2003; Macaulay et al. 2005; Atkinson et al. 2008). Lineages indigenous to Africa occur at the base of the mtDNA tree and are usually divided into four main haplogroups: L0 (formerly L1a, L1d, L1f and L1k); L1; L2; and L3 (figure 1). Four other haplogroups are sometimes identified (L4–L7), but these are relatively rare (Gonder et al. 2007). Haplogroup L0 is found in southern Africa and eastern/southeastern Africa (Salas et al. 2002; Gonder et al. 2007). Its oldest branches (L0d and L0k) occur at high frequencies only among the Khoisan hunter–gatherers of South Africa (Watson et al. 1997; Salas et al. 2002). Haplogroup L1 is found at moderately high frequencies in western and central sub-Saharan Africa, including in the pygmy populations of the central equatorial forest region (Salas et al. 2002). Haplogroup L2 is common in western and southeastern sub-Saharan Africa and is the most numerous and widespread of the four major haplogroups, accounting for as much as 25 per cent of indigenous haplotype variation (Salas et al. 2002). The youngest of the major haplogroups, L3, is most common in western and eastern/southeastern sub-Saharan Africa, particularly among speakers of the Bantu language family, and is thought to have originated in eastern Africa, where it accounts for half of all types (Salas et al. 2002). Two other haplogroups descended from L3 (M and N) are the only mtDNA lineages observed outside Africa.
Figure 1.
Genealogy of major African mtDNA haplogroups. This phylogeny shows the genealogical relationships between the L0, L1, L2 and L3 mtDNA lineages of Africa and the position of the two major non-African lineages, M and N.
The different temporal and geographical distributions of these four haplogroups make them potentially useful as markers of broad population demographic processes within Africa over the last 150 000 years. The oldest lineages, haplogroups L0 and L1, occur at higher frequencies among hunter–gatherers and may provide a window into our earliest history. For example, the occurrence of certain L0 haplotypes among the Khoisan has recently been shown to support a split between the ancestral Khoisan population and other lineages between 90 and 150 kyr ago (Behar et al. 2008). The younger L2 and L3 haplogroups are much more widespread and common, but it is unclear how they achieved their current distribution and why the L3 haplogroup was the only lineage to be carried out of Africa.
Information about ancestral population size and growth parameters can be inferred from extant genetic variation using coalescent theory (Kingman 1982; Hudson 1990; Griffiths & Tavaré 1994). Early coalescent estimates of past African population demographics were based on the distribution of pairwise sequence differences in restriction fragment length polymorphism (RFLP) and D-loop data, assumed a single exponentially growing population and included a large degree of statistical uncertainty (Harpending et al. 1993; Sherry et al. 1994; Harpending & Rogers 2000). More recent work using complete mtDNA sequences has focused on the geographical and phylogenetic relationships between specific haplotypes and their TMRCAs but has not explicitly modelled population size through time (Watson et al. 1997; Bandelt et al. 2001; Salas et al. 2002; Gonder et al. 2007). Watson et al. (1997) were able to use median networks to identify star-like patterns in L2 and L3 RFLP and D-loop sequence diversity, which they suggest reflect an expansion 60–80 kyr ago, possibly linked to the subsequent human expansion from Africa. However, without model-based coalescent inference, it is difficult to evaluate hypotheses about the dispersal of the different haplogroups. Inferred lineage coalescence events constitute a single realization of a stochastic process, meaning there is no simple correspondence between lineage TMRCAs and population expansions.
Improvements in the available data and inference methods mean that it is now possible to estimate population demographic parameters through time with increased accuracy, without assuming a simple parametric growth curve and with an explicit framework for quantifying uncertainty in population parameter estimates (Shapiro et al. 2004; Atkinson et al. 2008). Given a sample of sequences from a population, the Bayesian skyline plot (BSP; Drummond & Rambaut 2003; Drummond et al. 2005) uses Bayesian coalescent inference together with a Markov chain Monte Carlo (MCMC; Metropolis et al. 1953; Hastings 1970) sampling algorithm to provide a credibility interval for effective population size through time that incorporates uncertainty in the substitution model parameters, underlying genealogy and coalescent times. Unlike previous approaches to estimating population size, the BSP does not require a pre-specified parametric growth model, and, because the method is applied directly to a set of sequence data rather than to pairwise sequence differences, it avoids the loss of information that is associated with distance-based methods (Steel et al. 1988). Elsewhere (Atkinson et al. 2008), we have used BSPs together with a large dataset of complete human mtDNA sequences to estimate the relative size and timing of regional human population expansions around the globe. We showed that coalescent estimates of present-day effective population sizes from complete mtDNA sequence diversity were a good predictor of relative contemporary population sizes inferred from historical and anthropological data. Estimates of ancestral effective population size in Africa revealed slow, relatively constant growth, but because the continent was treated as a single population, we would not expect to detect the expansion of any subgroup of the population that did not substantially change the overall population size.
Here, we use BSPs together with a dataset of African complete mtDNA sequences to estimate the relative population size of the four major mtDNA haplogroups through time. These plots do not equate to enduring physically separated populations, which are difficult to identify in Africa, but rather represent the expansion of four separate mitochondrial lineages within the African meta-population. It is expected that population sizes will be strongly correlated further back in time, reflecting the small founding population that the lineages arose from. By plotting the lineages separately, we can answer questions about the timing of later expansions within Africa that may have been linked to populations carrying the different haplogroups. For example, do haplogroups L0 and L1 show significantly different demographic histories, perhaps suggesting population divisions among the earliest humans? When did L2, Africa's most common haplogroup, begin to spread and how quickly did this occur? And, perhaps most intriguing, when did the L3 lineage begin to spread in Africa and what can this tell us about our subsequent expansion from our homeland?
2. Material and methods
(a) Sequence data
We compiled a dataset of 224 complete African mtDNA sequences from those reported in the literature (Ingman et al. 2000; Maca-Meyer et al. 2001; Mishmar et al. 2003; Macaulay et al. 2005; Kivisild et al. 2006; Torroni et al. 2006; Gonder et al. 2007; see table S1 in the electronic supplementary material). Only sequences identified as L haplotypes were sampled, and these were classified as L0 (n=60), L1 (n=41), L2 (n=43) or L3 (n=80). We included L4 (also known as L3g) as part of L3 and ignored the rarer haplotypes L5–L7. Excluding L4 from the analysis did not notably affect the results for L3 (see fig. S1 in the electronic supplementary material). Sequences were machine aligned in ClustalX (Thompson et al. 1997) using reference sequence J01415.1. The coalescent inference method we used to analyse the data assumes that sequences are sampled at random from the population of interest. Here, because we are treating each haplogroup as a separate population, we avoided sampling sequences from studies that focused on particular subtypes within an African haplogroup. Of the remaining studies, designed to sample African mtDNA diversity, complete sequencing may be non-random due to patchy sampling and the desire to sequence representatives of rarer variants. We do not expect this to significantly affect our findings, because we are combining sequences from a number of independent studies and there is no reason to think that the sampling procedure would have systematically biased the representation of haplogroups across all of these studies. Repeating the analyses using subsets of the full dataset, in which sequences from the larger contributing studies (Kivisild et al. 2006; Gonder et al. 2007) were removed, did not affect our results, producing the same patterns that we report in §2b.
(b) Bayesian skyline plots
For each of the four L haplogroups, BSPs (Drummond et al. 2005) of effective population size through time were constructed using a MCMC (Metropolis et al. 1953; Hastings 1970) sampling algorithm, as implemented in BEAST v. 1.4 (Drummond & Rambaut 2007). We also constructed BSPs of sets of 100 sequences randomly sampled from the entire 224-sequence dataset to estimate total African population size through time. The underlying population size function of the BSP can be fitted using either a piecewise constant or a piecewise linear function of population size change. Here we report results based on a piecewise linear model made up of 10 control points. The qualitative results were not affected by the number of control points fitted or by using a piecewise constant function of effective population size. Effective population size is a compound population genetics parameter generally considered to be linearly proportional to census population size—here, the population of breeding females carrying haplotypes from each haplogroup. Other factors, such as local extinction with recolonization, selection and various forms of non-random mating (Wakeley 2000), can also influence effective population size estimates. As we discuss below, selective sweeps are unlikely to account for the patterns we observe and coalescent inference of population size from mtDNA diversity has been shown to be a good predictor of census population size in humans (Atkinson et al. 2008).
To infer the ancestral gene trees for each haplogroup, we used a general time-reversible (GTR) substitution model with site-specific rates for first, second and third codon positions. This model has been shown to fit the data better than the commonly used GTR+Γ+I model (Shapiro et al. 2006; Atkinson et al. 2008). Each MCMC sample was based on a run of 40 000 000 generations, sampled every 4000, with the first 4 000 000 generations discarded as burn-in. Examination of autocorrelation times of the MCMC plots indicated that runs had converged to the posterior distribution and provided an adequate sample. All runs had an effective sample size of at least 1000 for the parameters of interest.
In order to assign a time scale to the population size estimates, the rate of molecular evolution must be calibrated. Evidence for the existence of trends in observed substitution rates through time (Ho et al. 2005) makes an internal rate calibration (of similar time depth to the period of interest in Africa) preferable to the commonly applied human–chimpanzee calibration. Following previous work analysing a global sample of human mtDNA diversity (Atkinson et al. 2008), rates were fixed to 1.691×10−8 substitutions per site per year. In order to separate uncertainty in population size estimates from rate calibration error, uncertainty in the calibration time itself was not included in the analyses reported here. This allows for a more accurate comparison of the relative timing of growth patterns between the haplogroups. The rate calibration is based on a 45 kyr age for haplogroup Q in New Guinea and earliest archaeological evidence of human habitation on the island is 42–45 kyr ago (O'Connell & Allen 2004). Haplogroup Q occurs at high frequencies only in New Guinea (a closely related haplogroup, Q2, found in parts of Melanesia, also occurs at low frequencies in Micronesia and Polynesia, probably due to more recent gene flow; Friedlaender et al. 2005). Ho & Endicott (2008) have suggested an alternative calibration linking haplogroup P to the colonization of Sahul, which yields slightly faster rates if the same colonization time is assumed for Sahul. We note, however, that absolute time estimates scale in proportion to the chosen calibration age, such that if haplogroup Q is assumed to be 40 kyr old, all inferred dates would simply decrease by approximately 11 per cent.
3. Results and discussion
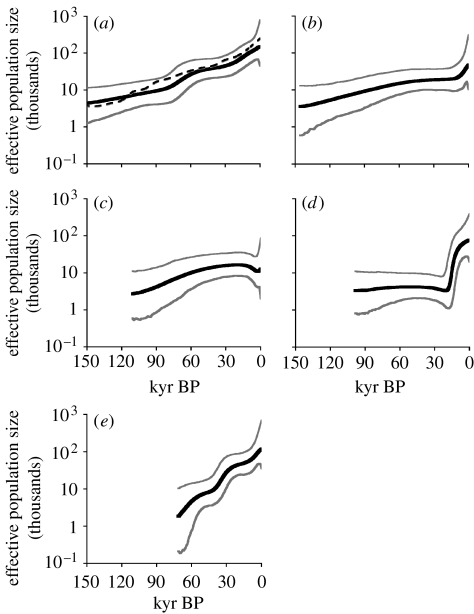
Figure 2 shows BSPs of effective population size through time for the total African population and for each of the four major haplogroups. The bold black line represents the median population size estimate from the Bayesian posterior distribution. The grey lines delimit the 95% highest posterior density (HPD) boundaries. Consistent with our previous findings using a smaller African sample (Atkinson et al. 2008), when considered as a single population, total sub-Saharan African effective population size estimates (figure 2a) show gradual, roughly exponential growth from a TMRCA 150–201 kyr ago (95% HPD)—with perhaps a slight increase in growth rate ca 70 kyr ago. As expected, the same gradual growth pattern is revealed when population size estimates are summed across the separate lineage analyses (dashed line, figure 2a). This pattern contrasts with evidence for rapid growth outside Africa (Atkinson et al. 2008) and is consistent with findings from independent genetic loci, including the X- and Y-chromosomes (Garrigan et al. 2007), and microsatellite and single-nucleotide polymorphism data (Gonser et al. 2000; Zhivotovsky et al. 2000; Alonso & Armour 2001; Rogers 2001; Marth et al. 2004). This broad agreement across multiple loci makes it likely that the growth signals we infer reflect population expansions rather than multiple simultaneous selective sweeps.
Figure 2.
BSPs of effective population size through time for (a) sub-Saharan Africa (n=224), (b) haplogroup L0 (n=60), (c) haplogroup L1 (n=41), (d) haplogroup L2 (n=43) and (e) haplogroup L3 (n=80). The bold black line represents the median posterior effective population size through time. The grey lines delimit the 95% highest posterior density for effective population size, accounting for uncertainty in the reconstructed phylogeny and substitution model parameters. The dashed line in (a) plots the sum of the separate lineage population estimates through time. Effective population size is plotted on a log scale and assumes a generation time of 20 years. These estimates of effective population size have an inverse relationship with the evolutionary rate of mtDNA used for the calibration, such that they will be lower for faster rates and higher for slower rates. For comparison, all the x-axes have a scale extending to 150 kyr. The plots are truncated to the median estimate of each region's TMRCA.
However, the separate BSPs for the four major haplogroups (figure 2b–e) reveal very different patterns of growth from the overall picture, particularly for haplogroups L2 and L3. Examination of the plots indicates that growth signals in haplogroups L0 (figure 2b) and L1 (figure 2c) are not significantly different from each other, showing substantial overlap in HPD distributions through time. However, HPD distributions for haplogroups L2 (figure 2d) and L3 (figure 2e) are significantly different from each other and from haplogroups L0 and L1. These differences may reflect changes in the size or structure of the populations carrying these haplotypes or could indicate the influence of selection pressures causing advantageous new variants to sweep through the population. We discuss the implications of the different growth profiles below.
Haplogroups L0 and L1 (figure 2b,c, respectively) show slow constant growth over the last 100–200 kyr (TMRCAs: L0, 124–172 kyr ago; L1, 87–139 kyr ago; L0 and L1 combined, 156–213 kyr ago; 95% HPDs). Estimates of effective population size do not differ significantly throughout the history of the two lineages. We would expect to see this pattern if both lineages formed part of an early panmictic African population and contributed relatively equally to the haplotype diversity present in Africa today. However, recent evidence indicates some population structure within Africa from an early stage. Analysis of genetic data from four African populations using a coalescent-based model of population divergence found support for population structure in Africa from more than 50 kyr ago when applied to mtDNA, although not when applied to Y-chromosome data (Garrigan et al. 2007). Others have argued that the distribution of ancient mtDNA L0d and L0k lineages among the hunter–gatherer speakers of the African ‘click’ languages also supports population structure deep in the mtDNA tree (Knight et al. 2003; Tishkoff et al. 2007; Behar et al. 2008). Our findings show that, whether such deep population structure exists or not, it did not produce substantially different population growth profiles between the most ancient lineages. Repeating the BSP analysis on only the L0d and L0k lineages also did not produce significantly different growth profile from the L0 BSP (see fig. S2 in the electronic supplementary material). This makes it less likely that such putative deep divergence events were associated with a substantial change in available territory or mode of living. Effective population size plots for the two haplogroups do appear to be on different trajectories over the last 15 kyr, although they do not differ significantly. L0 shows an increasing trend while L1 is decreasing. Inspection of the underlying phylogeny indicates that the increase in L0 is probably a result of a rapid emergence of lineages within the L0a haplogroup at this time. L0a has been proposed as a marker of the Bantu expansion 3000–4000 years ago (Bandelt et al. 1995; Chen et al. 1995). The time scale we reconstruct would suggest it began to expand earlier, perhaps linked to the expansion of L2a, discussed below.
For L2, we infer a TMRCA of 73–127 kyr ago (95% HPD). This range is consistent with previous age estimates for the haplogroup based on D-loop and complete sequence data and has led to the suggestion that L2 may have been involved in population expansions associated with the later African exodus (Watson et al. 1997; Salas et al. 2002). However, the BSP in figure 2d shows L2 to have been at relatively low frequency until a period of substantial growth beginning 12–20 kyr ago (95% HPD). This is the most pronounced expansion signal we observe among the major haplogroups and probably explains the high frequency of L2 lineages across Africa today. The most common haplogroup within L2 is L2a (TMRCA 25–31 kyr ago). Repeating the analysis on just the L2a lineages confirms that they are the principal source of the L2 expansion signal (see fig. S3 in the electronic supplementary material). While this rapid expansion signal in L2 could have been caused by a selective sweep, the substitutions that define the L2 (and L2a) lineages are synonymous changes, making it unlikely that these were new and advantageous variants capable of quickly displacing existing haplotypes. As with L0a mentioned above, L2a has been linked to the spread of Bantu languages (Bandelt et al. 2001). However, as with L0a, the timing of the growth we infer here pre-dates the Bantu expansion. This discrepancy could be explained as a result of the shift in inferred rates of mtDNA evolution at shallow time depths identified by Ho et al. (2005), although rates would need to be four to five times faster to bring the start of the growth phase seen in L2a into agreement with the proposed spread of Bantu 3000–4000 years ago. While the Bantu expansion undoubtedly played a role in the spread of a number of mtDNA haplotypes, L2 appears to have begun to expand somewhat earlier. Our findings fit with a proposal that L2 lineages (and perhaps the L0a lineages) spread as a result of environmental changes associated with the Last Glacial Maximum (Salas et al. 2002). Particularly arid conditions at this time are thought to have resulted in the enlargement of the Sahara as well as the conversion of much of the forested area of central Africa to open savannah and woodlands (Adams & Faure 1997), which may have allowed an expansion of human populations into new territories, particularly in central Africa.
Figure 2e shows the BSP for haplogroup L3 and reveals a marked increase in effective population size from an estimated TMRCA of 61–86 kyr ago (95% HPD). L3 is the only lineage to show such marked growth at this time. Again, the substitutions that define L3 are synonymous changes, hence the inferred increase in L3 frequency is unlikely to be the result of a selective sweep. Instead, it seems likely that L3 spread due to demographic expansion within Africa. L3 haplotype frequencies are highest in eastern Africa (Salas et al. 2002), the proposed launching point of the human colonization of the globe. The timing of the L3 expansion pre-dates the emergence of the first non-African lineages (haplogroups M and N) by 8–12 kyr—based on the same rate calibration used here, we estimate an age of 53–69 and 50–64 kyr ago (95% HPDs) for M and N, respectively (Atkinson et al. 2008). The fact that L3 is the only haplogroup with descendants outside Africa and shows a clear growth signal 8–12 kyr prior to the emergence of its non-African descendants strongly suggests that L3 did not simply spill over into Eurasia, but was driven as part of an expansion that had begun in sub-Saharan Africa thousands of years earlier.
There are a number of possible explanations for such an expansion. A recent study of drill cores from Lake Malawi sediment has indicated a change from highly variable arid conditions to a wetter, more stable climate in eastern Africa ca 70 kyr ago (Scholz et al. 2007). The authors argue that such a climatic shift could have stimulated human population growth and later migrations from the region. While we would find this argument persuasive, if climate was the only factor at work, the other haplogroups, also thought to be in eastern Africa at this time (Salas et al. 2002), should show a similar expansion pattern. We can explain the observed pattern of growth if an initially small group carrying the ancestral L3 haplotype gained a cultural advantage that allowed its population to out-compete rival groups. Consistent with this idea, Mellars (2006) has argued on the basis of archaeological evidence that the African exodus was pre-dated by a cultural revolution involving new stone blade technologies, skin working tools, ornaments and imported red ochre (although cf. Marean et al. 2007 for an earlier date). Red ochre, also present in the earliest known human remains from Australia (Bowler & Thorne 1976), is especially interesting in this context for its association with ritual and symbolism. More advanced symbolic systems in language and religious beliefs could have provided a competitive advantage to a group by promoting coordination and cohesion. Determining which, if any, of these factors were involved in the L3 expansion will require the synthesis of multiple lines of evidence from archaeology, palaeoclimatology, historical linguistics and population genetics. Future work in population genetics, extending coalescent methods such as those used here beyond a single locus to include Y-chromosome and genomic data, will further improve estimates of ancestral human population demographics and therefore help to provide an increasingly detailed understanding of the human story.
Acknowledgments
We acknowledge support from Leverhulme Trust grant F/00 239/T543 and the EC-FP6 EXREL project 43225.
Supplementary Material
Table S1, figures S1–S3
References
- Adams, J. M. & Faure, H. (eds) 1997 Review and atlas of palaeovegetation: preliminary land ecosystem maps of the world since the Last Glacial Maximum Oak Ridge, TN: Oak Ridge National Laboratory.
- Alonso S., Armour J.A. A highly variable segment of human subterminal 16p reveals a history of population growth for modern humans outside Africa. Proc. Natl Acad. Sci. USA. 2001;98:864–869. doi: 10.1073/pnas.98.3.864. doi:10.1073/pnas.011244998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson Q.D., Gray R.D., Drummond A.J. MtDNA variation predicts population size in humans and reveals a major southern Asian chapter in human prehistory. Mol. Biol. Evol. 2008;25:468–474. doi: 10.1093/molbev/msm277. doi:10.1093/molbev/msm277 [DOI] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Sykes B.C., Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.J., et al. Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade. Ann. Hum. Genet. 2001;65:549–563. doi: 10.1017/S0003480001008892. doi:10.1046/j.1469-1809.2001.6560549.x [DOI] [PubMed] [Google Scholar]
- Behar D., et al. The dawn of human matrilineal diversity. Am. J. Hum. Genet. 2008;82:1–11. doi: 10.1016/j.ajhg.2008.04.002. doi:10.1016/j.ajhg.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler J.M., Thorne A.G. Human remains from Lake Mungo: discovery and excavation of Lake Mungo III. In: Kirk R.L., Thorne A.G., editors. The origin of the Australians. Australian Institute of Aboriginal Studies; Canberra, Australia: 1976. pp. 127–138. [Google Scholar]
- Cann R.L., Stoneking M., Wilson A.C. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. doi:10.1038/325031a0 [DOI] [PubMed] [Google Scholar]
- Chen Y.S., Torroni A., Excoffier L., Santachiarabenerecetti A.S., Wallace D.C. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am. J. Hum. Genet. 1995;57:133–149. [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2003 Beast, v. 1.3. Oxford, UK: Oxford University Press. (http://evolve.zoc.ox.ac.uk/beast/)
- Drummond A.J., Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. doi:10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A., Shapiro B., Pybus O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. doi:10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- Forster P., Matsumura S. Evolution. Did early humans go north or south? Science. 2005;308:965–966. doi: 10.1126/science.1113261. doi:10.1126/science.1113261 [DOI] [PubMed] [Google Scholar]
- Forster P., Harding R., Torroni A., Bandelt H.J. Origin and evolution of native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Friedlaender J., et al. Expanding southwest Pacific mitochondrial haplogroups P and Q. Mol. Biol. Evol. 2005;22:1506–1517. doi: 10.1093/molbev/msi142. doi:10.1093/molbev/msi142 [DOI] [PubMed] [Google Scholar]
- Garrigan D., et al. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. doi:10.1534/genetics.107.077495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder M.K., Mortensen H.M., Reed F.A., de Sousa A., Tishkoff S.A. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol. Biol. Evol. 2007;24:757–768. doi: 10.1093/molbev/msl209. doi:10.1093/molbev/msl209 [DOI] [PubMed] [Google Scholar]
- Gonser R., Donnelly P., Nicholson G., Di Rienzo A. Microsatellite mutations and inferences about human demography. Genetics. 2000;154:1793–1807. doi: 10.1093/genetics/154.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R.C., Tavaré S. Simulating probability distributions in the coalescent. Theor. Popul. Biol. 1994;46:131–159. doi:10.1006/tpbi.1994.1023 [Google Scholar]
- Harpending H., Rogers A. Genetic perspectives on human origins and differentiation. Annu. Rev. Genom. Hum. Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. doi:10.1146/annurev.genom.1.1.361 [DOI] [PubMed] [Google Scholar]
- Harpending H.C., Sherry S.T., Rogers A.R., Stoneking M. The genetic structure of ancient human populations. Curr. Anthropol. 1993;34:483–496. doi:10.1086/204195 [Google Scholar]
- Hastings W.K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. doi:10.1093/biomet/57.1.97 [Google Scholar]
- Ho S.Y., Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am. J. Hum. Genet. 2008;83:142–146. doi: 10.1016/j.ajhg.2008.06.014. doi:10.1016/j.ajhg.2008.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.Y., Phillips M.J., Cooper A., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. doi:10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Hudson R.R. Gene genealogies and the coalescent process. Oxf. Surv. Evol. Biol. 1990;7:1–14. [Google Scholar]
- Ingman M., Kaessmann H., Paabo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. doi:10.1038/35047064 [DOI] [PubMed] [Google Scholar]
- Kingman J.F.C. On the genealogy of large populations. J. Appl. Probab. 1982;19A:27–43. doi:10.2307/3213548 [Google Scholar]
- Kivisild T., et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. doi:10.1534/genetics.105.043901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A., Underhill P.A., Mortensen H.M., Zhivotovsky L.A., Lin A.A., Henn B.M., Louis D., Ruhlen M., Mountain J.L. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr. Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. doi:10.1016/S0960-9822(03)00130-1 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N., Gonzalez A.M., Larruga J.M., Flores C., Cabrera V.M. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001;2:13. doi: 10.1186/1471-2156-2-13. doi:10.1186/1471-2156-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V., et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. doi:10.1126/science.1109792 [DOI] [PubMed] [Google Scholar]
- Marean C.W., et al. Early human use of marine resources and pigment in South Africa during the Middle Pleistocene. Nature. 2007;449:905–908. doi: 10.1038/nature06204. doi:10.1038/nature06204 [DOI] [PubMed] [Google Scholar]
- Marth G.T., Czabarka E., Murvai J., Sherry S.T. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. doi:10.1534/genetics.166.1.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc. Natl Acad. Sci. USA. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. doi:10.1073/pnas.0510792103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis N., Rosenbluth A.W., Rosenbluth M.N., Teller A.H., Teller E. Equations of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1091. doi:10.1063/1.1699114 [Google Scholar]
- Mishmar D., et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. doi:10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell J.F., Allen J. Dating the colonization of Sahul (Pleistocene Australia–New Guinea): a review of recent research. J. Archaeol. Sci. 2004;31:835–853. doi:10.1016/j.jas.2003.11.005 [Google Scholar]
- Olivieri A., et al. The mtDNA legacy of the Levantine Early Upper Palaeolithic in Africa. Science. 2006;314:1767–1770. doi: 10.1126/science.1135566. doi:10.1126/science.1135566 [DOI] [PubMed] [Google Scholar]
- Rogers A.R. Order emerging from chaos in human evolutionary genetics. Proc. Natl Acad. Sci. USA. 2001;98:779–780. doi: 10.1073/pnas.98.3.779. doi:10.1073/pnas.98.3.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A., Richards M., De la Fe T., Lareu M.V., Sobrino B., Sanchez-Diz P., Macaulay V., Carracedo A. The making of the African mtDNA landscape. Am. J. Hum. Genet. 2002;71:1082–1111. doi: 10.1086/344348. doi:10.1086/344348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C.A., et al. East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc. Natl Acad. Sci. USA. 2007;104:16 416–16 421. doi: 10.1073/pnas.0703874104. doi:10.1073/pnas.0703874104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B., et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. doi:10.1126/science.1101074 [DOI] [PubMed] [Google Scholar]
- Shapiro B., Rambaut A., Drummond A.J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. doi:10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- Sherry S.T., Rogers A.R., Harpending H., Soodyall H., Jenkins T., Stoneking M. Mismatch distributions of mtDNA reveal recent human population expansions. Hum. Biol. 1994;66:761–775. [PubMed] [Google Scholar]
- Steel M.A., Hendy M.D., Penny D. Loss of information in genetic distances. Nature. 1988;336:118. doi: 10.1038/336118a0. doi:10.1038/336118a0 [DOI] [PubMed] [Google Scholar]
- Thangaraj K., Chaubey G., Kivisild T., Reddy A.G., Singh V.K., Rasalkar A.A., Singh L. Reconstructing the origin of Andaman islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. doi:10.1126/science.1109987 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A., et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. doi:10.1093/molbev/msm155 [DOI] [PubMed] [Google Scholar]
- Torroni A., Achilli A., Macaulay V., Richards M., Bandelt H.J. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. doi:10.1016/j.tig.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Underhill P.A., Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. doi:10.1146/annurev.genet.41.110306.130407 [DOI] [PubMed] [Google Scholar]
- Underhill P.A., et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. doi:10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A.C. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. doi:10.1126/science.1840702 [DOI] [PubMed] [Google Scholar]
- Wakeley J. The effects of subdivision on the genetic divergence of populations and species. Evol. Int. J. Org. Evol. 2000;54:1092–1101. doi: 10.1111/j.0014-3820.2000.tb00545.x. doi:10.1111/j.0014-3820.2000.tb00545-x [DOI] [PubMed] [Google Scholar]
- Watson E., Forster P., Richards M., Bandelt H.J. Mitochondrial footprints of human expansions in Africa. Am. J. Hum. Genet. 1997;61:691–704. doi: 10.1086/515503. doi:10.1086/515503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon D.J., et al. Human population genetic structure and diversity inferred from polymorphic L1 (LINE-1) and Alu insertions. Hum. Hered. 2006;62:30–46. doi: 10.1159/000095851. doi:10.1159/000095851 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L.A., Bennett L., Bowcock A.M., Feldman M.W. Human population expansion and microsatellite variation. Mol. Biol. Evol. 2000;17:757–767. doi: 10.1093/oxfordjournals.molbev.a026354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, figures S1–S3