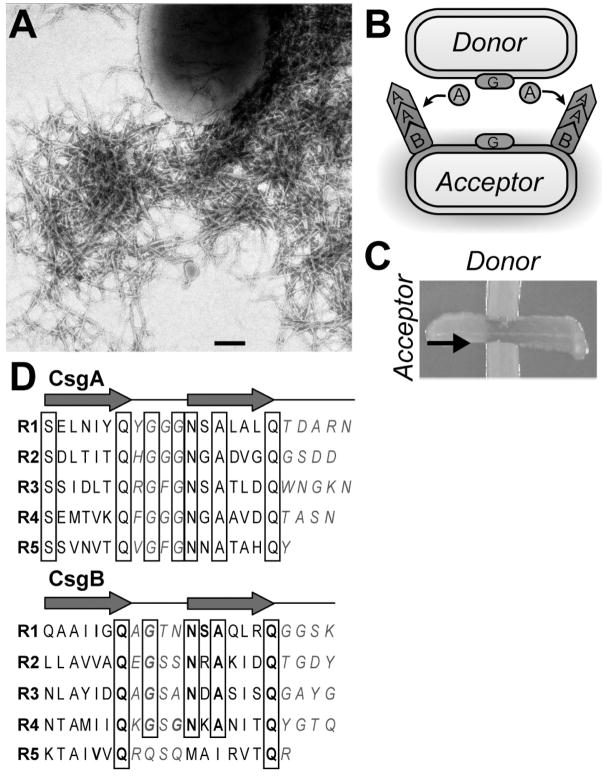
Fig. 2.
Interaction between the curli subunit proteins CsgA and CsgB. (A) Negative stain electron micrograph of wild type cells producing curli. Scale bar represents 200 nanometers. (B) Model of Interbacterial Complementation. A donor cells secretes soluble CsgA that acts as a substrate for CsgB on an acceptor cells where curli biogenesis takes place. (C) A Congo red indicator plate demonstrating interbacterial complementation. The donor cells and the acceptor cells appear white until the two strains intersect. Once the two cell types intersect Congo red binding occurs demonstrating curli fiber polymerization as taken place. The arrow represents the direction in which the acceptor cells were streaked onto the plate. (D) The oligopeptide repeating units that compose the CsgA and CsgB proteins. The three dimensional structures of CsgA and CsgB are predicted to be composed of five imperfect β-strand-loop-β-strand oligopeptide repeats (R1–R5). Amino acids comprising the β-strand are located below the arrows, and amino acids predicted to comprise the loops are denoted with italicized blue letters. Bolded letters represent amino acids conserved in CsgB and CsgA at each position relative to the start of each repeating unit. Boxed letters represent amino acids conserved throughout the repeating units in both proteins.