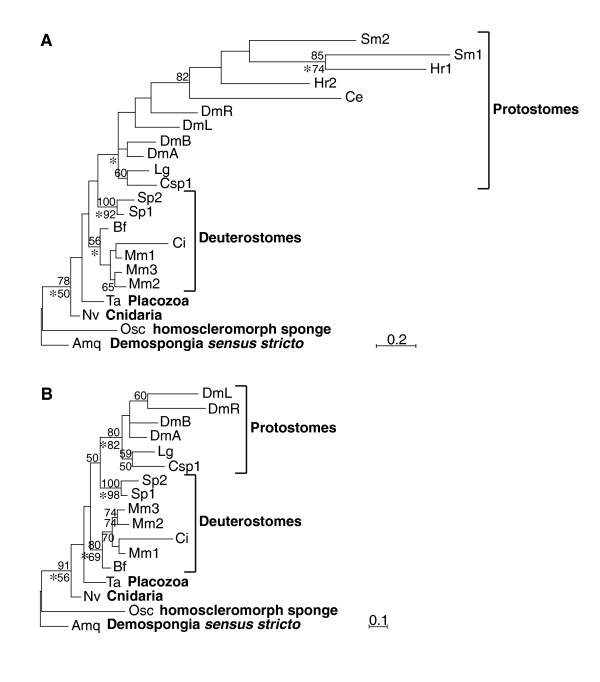
Figure 2.
Bayesian trees of Runx sequences. In a first analysis (A), all the genes from Figure 1 were included and, in a second analysis (B), long-branched taxa (Runx genes from S. mediterranea, H. robusta, and C. elegans) were excluded from the dataset. The trees were calculated using a multiple sequence alignment of amino acid sequences corresponding to the Runt domain of each species. Percentages of bootstrap support greater than or equal to 50% are indicated above the node for the distance analysis (Phylip 3.6; 1000 replicates) and below the node for the maximum likelihood analysis (Phylip 3.6; 100 replicates). An asterisk under the node indicates a Bayesian posterior probability greater than or equal to 95%. Abbreviations as in Figure 1.