Figure 1. A schematic representation of all of the mutations affecting the ZIC2 coding region sorted by their position and category.
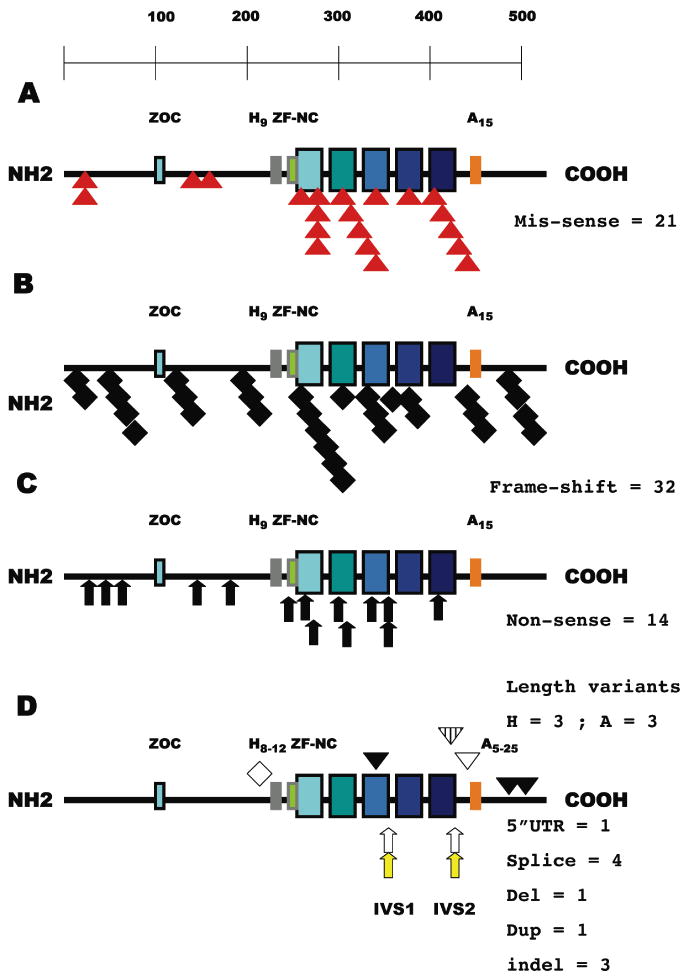
A: Mis-sense variations (red triangles, 25% overall) cluster within the zinc-finger motifs (17/21 = 81%). The ZIC2 transcription factor is illustrated as a line of aminoacids (from left to right, NH2 to COOH) with key motifs (Aruga et al., 2004, 2006) depicted by rectangular boxes: ZOC homology block (blue), His-9 variable region (grey, range 8-12), homology block ZF-NC (green) preceding the first of five zinc-fingers (increasing shades of blue to indicate that each finger has a unique structure) and Ala-15 variable region (orange, range 5-25). B: 32/83 = 38.5% of the mutations (black diamond) would predict a significant alteration of the reading frame and variable truncations primarily affecting DNA binding. C: Note that all non-sense truncations (black arrows, 17% overall) occur prior to zinc-fingers 4 and 5 primarily implicated in DNA binding by Krüppel/GLI-related transcription factors (Kinzler et al., 1990; Pavletich and Pabo, 1993). D: The typical observed range of poly-Alanine and poly-Histidine segments are shown (Brown et al., 1998, 2001, 2002; Dubourg et al., 2004). All four invariant splice sites including both donor (arrow, no fill) and both acceptor (yellow arrow) sites are subject to mutation. Note that the failure to initiate a splice from IVS2 donor site nevertheless maintains the reading frame predicting a protein with 71 additional aminoacids and a distortion of the zinc-finger 5 (inverted hatched triangle, see also Figure 2). Duplications that retain or resume frame are shown as diamonds with no fill while the inverted open triangle illustrates an in frame deletion. Complex insertion/deletion sequence variants causing a shift of the reading frame are the black inverted triangles. The non-coding 5′ UTR variation is not indicated by any symbol.
