Figure 2. Detailed structure of the zinc-finger region.
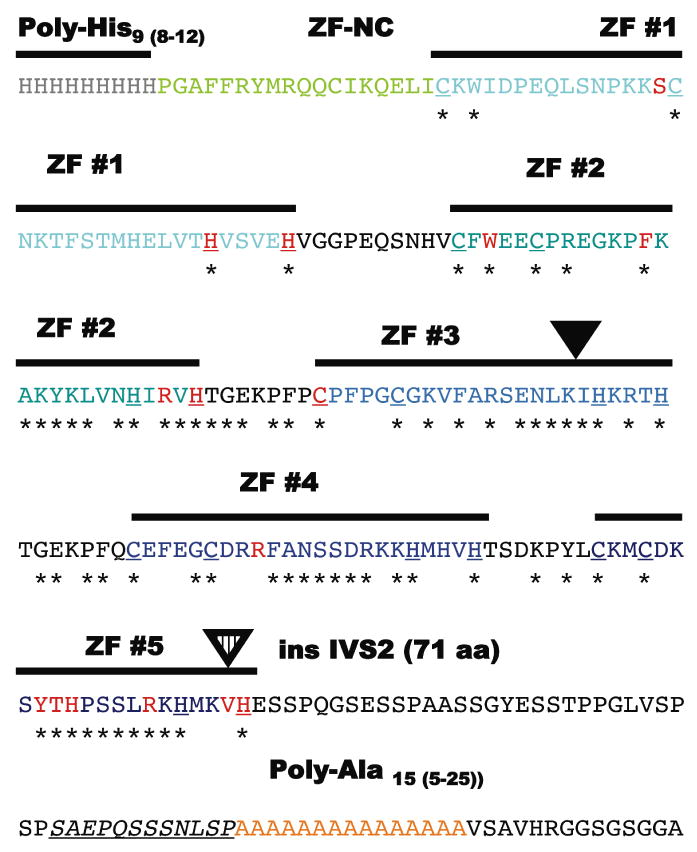
The aminoacid sequence (single aminoacid code) of the region from approximately His-9 to Ala-15 that contains the individually numbered Zinc-finger (ZF domains) and the most mutations with individual motifs colored as in Figure 1. The C2H2 components of each zinc-finger are bold and underlined and residues invariably present in the ZIC-family alignment are identified with an asterix. Note the non-random clustering of the mis-sense mutations (red) within these conserved aminoacid residues and particularly those that define the zinc-binding motif of the C2H2 zinc fingers (Redemann et al., 1988). Similarly, the addition of 7 novel aminoacids within the ZF #3 likely results in distorted alignment (inverted black triangle). The c.1239+1G>C (or IVS2+1G>C) allows for the incorporation in toto of the IVS2 into the reading frame of the transcript and adding an additional 71 aminoacids. This insertion also removes the ValHis sequence from its normal context within the C2H2 motif of ZF #5 and distorting its structure; this same key C2H2 Histidine is a target of mutation in another HPE individual.
