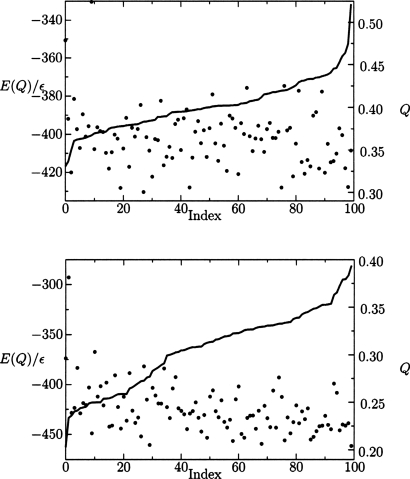
Figure 5.
The lowest energy structures of the training set protein, 434 repressor (top), and the blind prediction proteins, HDEA (bottom) identified from 100 independent basin-hopping simulations. Each minimum has values for energy, illustrated by dots, and structural overlap with the native state Q, represented by the continuous line. These minima are ordered with respect to their structural overlap Q with the native state (index). The data show correlations between the energy and Q, while the number of high quality structures is superior for the training protein.