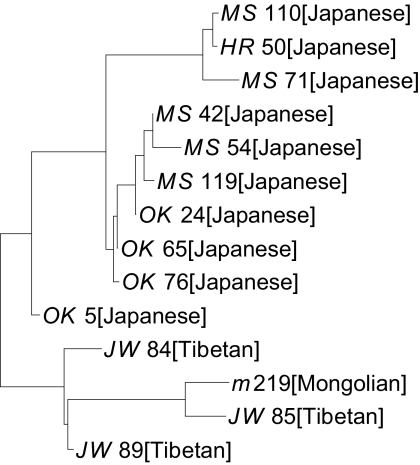
Figure 2.
Phylogenetic tree of haplogroup 4. Each allele is given by its identifier and its population of origin. We compared the MSY1 alleles of this haplogroup pairwise with our algorithm, MS_Align(version 2), and used the resulting pairwise distance matrix to infer an evolutionary tree for the alleles using a Neighbor-Joining phylogenetic reconstruction program, BIONJ [Gascuel, 1997]. This tree perfectly separates 10 Japaneses from 3 Tibetans and 1 Mongolian. The Japanese haplotypes coalesce in a single clade.