TABLE 2.
Linkage disequilibrium variance components within and among patients
| Sites | 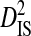 |
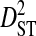 |
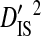 |
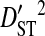 |
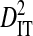 |
|---|---|---|---|---|---|
| 10, 32 | 0.00704 | 0.32857 | 0.33299 | 0.00144 | 0.33443 |
| 11, 13 | 0.00664 | 0.35998 | 0.36786 | 0.00506 | 0.37292 |
| 11, 25 | 0.00571 | 0.36850 | 0.37848 | 0.00109 | 0.37957 |
| 13, 14 | 0.00492 | 0.33667 | 0.32644 | 0.01206 | 0.33850 |
| 13, 25 | 0.00925 | 0.33989 | 0.35016 | 0.00342 | 0.35358 |
| 14, 22 | 0.00291 | 0.43311 | 0.42846 | 0.00548 | 0.43394 |
| 14, 25 | 0.00427 | 0.37203 | 0.38222 | 0.00221 | 0.38443 |
| 22, 25 | 0.00819 | 0.45563 | 0.46276 | 0.00458 | 0.46735 |
| 29, 32 | 0.00349 | 0.27981 | 0.28444 | 0.00264 | 0.28708 |
| 32, 34 | 0.00858 | 0.32199 | 0.32142 | 0.00075 | 0.32217 |
Sites shown are those with statistically significant linkage disequilibrium in both a data set containing one sequence from each of 3297 patients and a data set containing at least 100 sequences from each of 51 patients (8600 sequences in total). Variance components were estimated from the 51-patients data set.
