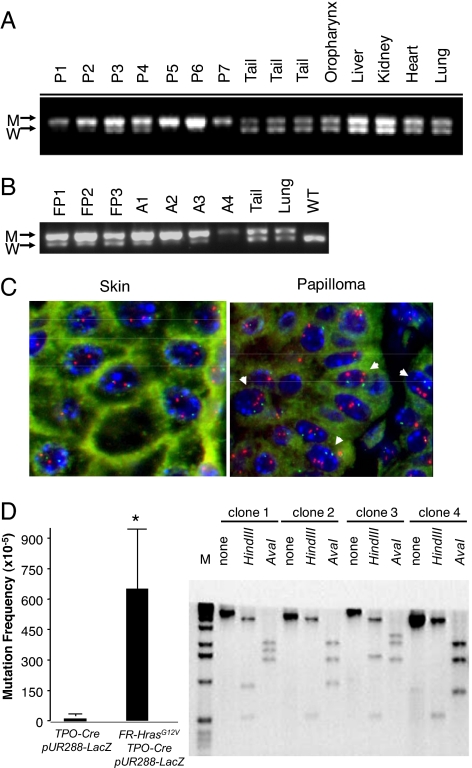
Fig. 4.
Hras allelic imbalance in papillomas and angiosarcoma from CC/FR-HrasG12V mice. (A) PCR of genomic DNA of papillomas with primers that distinguish mutant from WT Hras alleles (see Fig. 1A). W: 622 bp WT allele; M: 666 bp targeted allele because of insertion of loxP site. P1-P7: DNA from papillomas, or indicated nontumoral tissues. (B) PCR of DNA from forestomach papillomas (FP1–FP3), angiosarcomas (A1–A4), or indicated nontumoral tissues. (C) FISH of a representative section from papilloma tissue (P2) using a mouse BAC containing the Hras gene. Papilloma nuclei have 3 to 5 fluorescent signals corresponding to Hras (red), whereas adjacent skin is diploid. Mouse chromosome 7 centromeres are labeled in green. (D) (Left) Increased mutation rate in thyroid cells from FR-HrasG12V/TPO-Cre mice. Plasmids rescued from DNA extracts of thyroid glands of TPO-Cre/pUR288-LacZ or FR-HrasG12V/TPO-Cre/pUR288-LacZ were screened for alterations in LacZ, as described in Methods. Bars represent the mean mutation frequency ± SE of pooled samples from TPO-Cre/pUR288-LacZ (n = 3; 10 thyroids per pool) and FR-HrasG12V/TPO-Cre/pUR288-LacZ (n = 4; 10 thyroids per pool). P < 0.05. (Right) Representative Southern blot of LacZ-negative clones. The type of mutation was determined by PCR amplification and restriction digestion of LacZ-negative clones. Clone 1 contained an inactivating point mutation of LacZ, whereas the other 3 show distinct restriction profiles consistent with recombination events, insertions, or deletions.