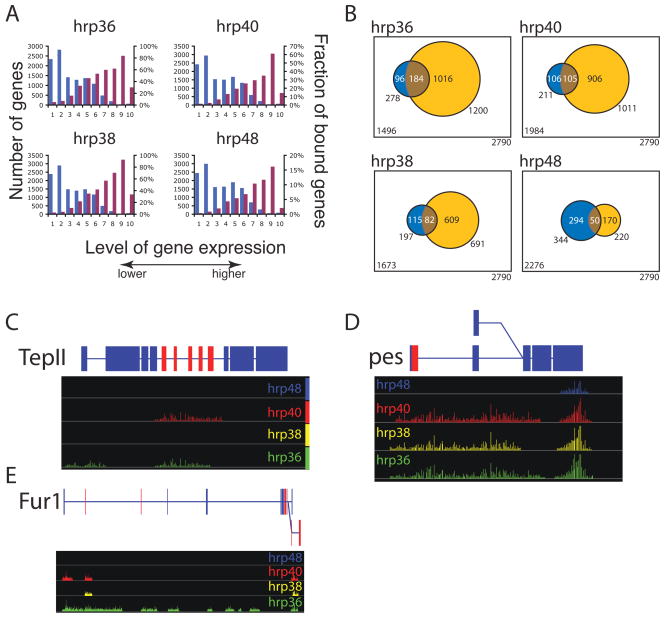
Figure 3.
Genome-wide identification of the RNAs associated with the different hnRNP proteins in RNP complexes. A) Distribution of the level of gene expression measured by Affymetrix arrays measured in S2 cells (blue bar) and the fraction of RNA bound by a hnRNP protein within a given level of expression (purple bar). B) The overlap between the genes that were found regulated in the splice junction microarray (blue) and bound in the tiling array (yellow). The highly significant overlapping transcripts are represented in brown (Fisher exact test, p-value<2×10−5). D) TepII and pes, two representative genes bound by hnRNP proteins showing extensive coverage suggestive of the spreading model. C) Fur1 a representative gene bound by hnRNP proteins showing discrete intronic binding tracts for hrp40 suggestive of the looping model. Significantly over-represented signal in the IP samples over the signal measured from RNA extracted from the starting RNP extract are represented by bars along the genome with the eight of the bar corresponding to p-values (expressed as −Log (p)). Above is a representation of the gene structure, blue boxes are constitutive exons, red boxes are alternative exons, lines are introns. Pes has two alternative promoters while Fur1 as two alternative poly-A signals represented by the 2 levels in the gene structure.