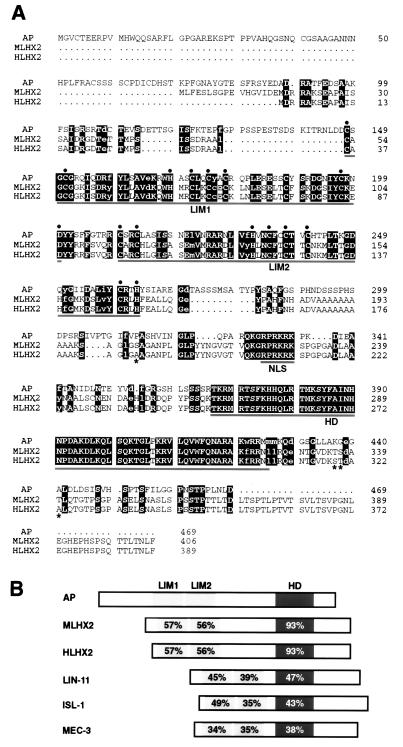
Figure 1.
Amino acid sequence comparison of Drosophila Ap and its mouse (MLHX2) and human (HLHX2) orthologs. (A) Sequence alignment. Identical amino acids between the three proteins are displayed in reverse type with capital letters, whereas conservative substitions are displayed in lower case letters. Asterisks indicate the only four different residues between the mouse and human proteins in the overlapping region. The two tandem LIM domains, the putative nuclear localization signal (NLS), and the homeodomain (HD) are underlined. The consensus residues of the LIM domains are highlighted with black circles. Gaps denoted by dots have been inserted to maximize sequence alignment. (B) Domain comparisons between Drosophila Ap, its mammalian orthologs, and other LIM-homeodomain proteins. These are: ISL-1 from rat (60), LIN-11 and MEC-3 from C. elegans (20, 21). Percentage of amino acid sequence identity is indicated within the LIM domains and homeodomain.