Table 1. Data-collection and refinement statistics for E2-25K M172A mutant structures.
Data were collected on SER-CAT beamline ID22. Values in parentheses are for the highest resolution shell.
| Low-pH crystal (PDB code 3e46) | High-pH crystal (PDB code 3f92) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.0 | 0.97 |
| Space group | I4 | I4 |
| Molecules per ASU | 1 | 1 |
| Unit-cell parameters (Å, °) | a = b = 134.5, c = 38.4, α = β = γ = 90 | a = b = 134.8, c = 38.2, α = β = γ = 90 |
| Resolution (Å) | 50.0–1.86 (1.91–1.86) | 50.0–2.23 (2.31–2.23) |
| Rmerge† (%) | 8.1 (37.4) | 6.9 (37.7) |
| 〈I/σ(I)〉 | 19.65 (1.98) | 11.95 (2.39) |
| Completeness (%) | 93.4 (52.4) | 98.2 (88.8) |
| No. of reflections (No. unique) | 109109 (27414) | 48847 (16794) |
| Redundancy | 4.0 (1.4) | 2.9 (1.9) |
| No. of frames | 125 | 75 |
| Matthews coefficient (Å3 Da−1) | 3.1 | 3.1 |
| Refinement | ||
| Resolution (Å) | 32.38-1.86 | 42.64-2.23 |
| No. of reflections | 27404 | 15939 |
| Rwork/Rfree‡ (%) | 17.4/21.0 | 17.4/21.3 |
| No. of atoms/molecules | ||
| Protein | 1597 | 1599 |
| Non-water molecules | 1 | 5 |
| Water molecules | 207 | 89 |
| R.m.s.d. bonds (Å) | 0.018 | 0.024 |
| R.m.s.d. angles (°) | 1.5 | 1.85 |
| Ramachandran plot | ||
| Most favored region (%) | 99.0 | 99.0 |
| Additional allowed region (%) | 1.0 | 1.0 |
R
merge = 
 , where I
i(hkl) is the observed intensity of reflection i and 〈I(hkl)〉 is the average intensity of multiple observations.
, where I
i(hkl) is the observed intensity of reflection i and 〈I(hkl)〉 is the average intensity of multiple observations.
R
work and R
free = 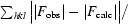
 , where R
free was calculated using 5% of the total reflections chosen at random and not used in the refinement.
, where R
free was calculated using 5% of the total reflections chosen at random and not used in the refinement.
