Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell (2.12.21).
| Crystal parameters | |
| Space group | P41212 |
| Unit-cell parameters (, ) | a = 57.69, b = 57.69, c = 172.60, = 90, = 90, = 90 |
| Data collection | |
| Resolution () | 47.962.10 (2.212.10) |
| Wavelength () | 0.9763 |
| R merge † (%) | 10.3 (50.6) |
| R p.i.m. ‡ (%) | 4.3 (20.5) |
| I/(I) | 13.5 (4.0) |
| No. of unique reflections | 17928 |
| Multiplicity | 6.7 (7.0) |
| Completeness (%) | 100 (100) |
| Refinement | |
| Resolution () | 2.1 |
| R § (%) | 0.206 |
| R free ¶ (%) | 0.241 |
| R.m.s.d. stereochemistry†† | |
| Bond lengths () | 0.114 |
| Bond angles () | 1.294 |
| No. of protein atoms | 1857 |
| No. of water molecules | 107 |
| Average B factor (2) | 18.09 |
| Ramachandran analysis‡‡ (%) | |
| Favoured | 96.2 |
| Allowed | 100.0 |
| Disallowed | 0 |
R
merge = 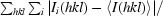
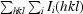 , where I
i(hkl) is the integrated intensity of a given reflection and I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections.
, where I
i(hkl) is the integrated intensity of a given reflection and I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections.
R
p.i.m. = 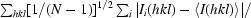
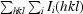 , where I
i(hkl) is the integrated intensity of a given reflection, I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections and N is the multiplicity of a given reflection.
, where I
i(hkl) is the integrated intensity of a given reflection, I(hkl) is the mean intensity of multiple corresponding symmetry-related reflections and N is the multiplicity of a given reflection.
R = 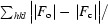
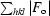 , where F
o and F
c are the observed and calculated structure factors, respectively.
, where F
o and F
c are the observed and calculated structure factors, respectively.
R free is the same as R but calculated using 5% random data excluded from the refinement.
R.m.s.d. stereochemistry is the deviation from ideal values.
Ramachandran analysis was carried out using MolProbity (Davis et al., 2007 ▶).
