Abstract
Small Heterodimer Partner (SHP) inhibits activities of numerous transcription factors involved in diverse biological pathways. As an important metabolic regulator, SHP plays a key role in maintaining cholesterol and bile acid homeostasis by inhibiting cholesterol conversion to bile acids. While SHP gene induction by increased bile acids is well established, whether SHP activity is also modulated remains unknown. Here, we report surprising findings that SHP is a rapidly degraded protein via the ubiquitin–proteasomal pathway and that bile acids or bile acid-induced intestinal fibroblast growth factor 19 (FGF19) increases stability of hepatic SHP by inhibiting proteasomal degradation in an extracellular signal-regulated kinase (ERK)-dependent manner. SHP was ubiquitinated at Lys122 and Lys123, and mutation of these sites altered its stability and repression activity. Tandem mass spectrometry revealed that upon bile acid treatment, SHP was phosphorylated at Ser26, within an ERK motif in SHP, and mutation of this site dramatically abolished SHP stability. Surprisingly, SHP stability was abnormally elevated in ob/ob mice and diet-induced obese mice. These results demonstrate an important role for regulation of SHP stability in bile acid signaling in normal conditions, and that abnormal stabilization of SHP may be associated with metabolic disorders, including obesity and diabetes.
Keywords: ERK, FGF19, SHP, bile acids, proteasome, protein stability
Cholesterol is a component of the cell membrane and is an essential precursor for steroid hormones, fat-soluble vitamins, and bile acids (Russell and Setchell 1992; Russell 1999). Bile acids play dietary roles in the absorption of lipid-soluble nutrients and also function as signaling molecules that coordinately regulate lipid and glucose metabolism and energy homeostasis (Russell 1999; Houten et al. 2006). Despite their essential functions, excess levels of cholesterol and bile acids cause metabolic diseases, such as cardiovascular disease, gall stone formation, hepatic cholestasis, and cancer. Therefore, cholesterol and bile acid levels must be tightly regulated. Indeed, the orphan nuclear receptor and transcriptional corepressor Small Heterodimer Partner (SHP) plays a key role in this regulation by inhibiting the biosynthesis of bile acids from cholesterol (Goodwin et al. 2000; Lu et al. 2000; Bavner et al. 2005).
Increased levels of bile acids feedback-inhibit transcription of the cholesterol 7α hydroxylase (CYP7A1) gene, which encodes the first and rate-limiting enzyme in hepatic conversion of cholesterol to bile acids (Jelinek et al. 1990; Chiang 2002). It is now apparent that feedback inhibition of CYP7A1 expression is a complicated process that involves multiple bile acid-activated signaling pathways, such as the FXR/SHP nuclear receptor cascade, cellular kinases, and the recently identified fibroblast growth factor 15/19 (FGF15/19) (Goodwin et al. 2000; Lu et al. 2000; Gupta et al. 2001; Kerr et al. 2002; Wang et al. 2002; Holt et al. 2003; Inagaki et al. 2005). SHP has been implicated as a key downstream regulator in all of these inhibitory pathways.
In the FXR/SHP pathway, the nuclear bile acid receptor FXR senses the elevated hepatic bile acid levels and induces SHP gene expression (Goodwin et al. 2000; Lu et al. 2000). Induced SHP inhibits transcription of CYP7A1 by coordinately recruiting the chromatin-modifying cofactors, mSin3A/HDAC corepressors, G9a methyltransferase, and Swi/Snf–Brm chromatin remodeling complex to the promoter (Kemper et al. 2004; Fang et al. 2007). The physiological relevance of SHP has been demonstrated by the observations that CYP7A1 expression and the bile acid pool size are increased in SHP-null mice (Kerr et al. 2002; Wang et al. 2002), whereas in transgenic mice overexpressing SHP in the liver, CYP7A1 expression and the bile acid pool size was decreased (Boulias et al. 2005). In addition to the FXR pathway, bile acids have been shown to inhibit CYP7A1 by activating cellular kinase pathways, such as c-jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and PKB (AKT) kinases (Gupta et al. 2001; Wang et al. 2002; Holt et al. 2003; Dent et al. 2005).
Bile acid-activated FXR was demonstrated recently to induce expression of FGF15/19 (FGF15 is the mouse homolog of human FGF19) in the small intestine. Secreted FGF15 inhibits CYP7A1 transcription in the liver by activating the membrane FGF receptor 4 (Inagaki et al. 2005). The inhibition of CYP7A1 by FGF15 was substantially reversed in SHP-null mice, indicating a critical role for SHP in FGF15 action. However, FGF15 treatment did not result in increased SHP mRNA levels in the liver, so the mechanism by which FGF15 acts in a SHP-dependent manner remains unknown, but could involve post-transcriptional mechanisms.
Post-translational modifications, such as ubiquitination and phosphorylation, dynamically regulate the activity and stability of transcriptional regulatory proteins. The ubiquitin–proteasome pathway is a major system for the degradation of short-lived regulatory proteins whose activities must be tightly regulated, such as NFκB, jun/fos, and p53 (Brooks and Gu 2003). Protein phosphorylation plays a critical role in regulating protein ubiquitination (Pickart 2001; Sundqvist et al. 2005). Since a role for the JNK, ERK, and p38 MAP kinase pathways in bile acid regulation of CYP7A1 has been demonstrated (Gupta et al. 2001; Holt et al. 2003; Dent et al. 2005; Xu et al. 2007), it is possible that phosphorylation of SHP mediated by these kinase signaling pathways could affect ubiquitination, and thereby regulate protein degradation of SHP.
In this study, we sought to determine whether bile acids and bile acid-induced intestinal FGF19 might increase hepatic SHP activity by increasing protein stability through post-translational modifications of SHP. From studies in vivo in mice, as well as in cultured hepatoma cells and primary human hepatocytes (PHH), we obtained compelling evidence indicating that SHP is rapidly degraded, with a half-life shorter than 30 min, via the ubiquitin–proteasomal pathway. Treatment with bile acids or FGF19 substantially increased SHP stability by inhibiting ubiquitination in an ERK-dependent manner. Surprisingly, hepatic SHP stability was abnormally elevated in ob/ob mice and mice fed a chronic western-style diet, implying a potential link between abnormal regulation of SHP stability and metabolic disorders.
Results
SHP is a target of ubiquitin–proteasomal degradation.
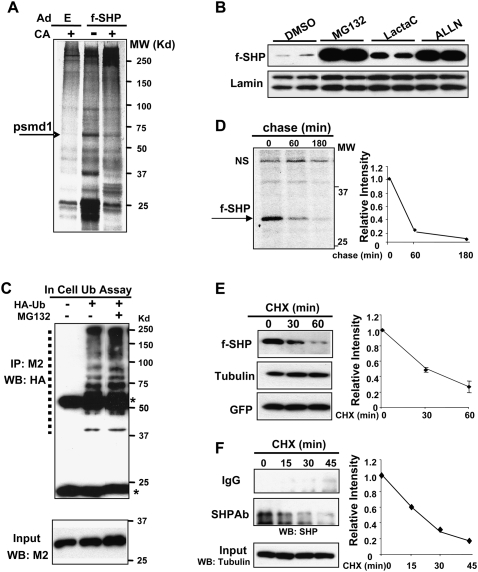
To identify novel SHP-interacting proteins that are involved in bile acid signaling in vivo, mice infected with adenoviral vectors expressing Flag-SHP were fed normal chow or cholic acid (CA)-supplemented chow. Hepatic proteins associated with Flag-SHP were isolated by M2 agarose, separated by SDS-PAGE, and identified by tandem mass spectrometry (MS/MS). Interaction of a 65-kDa protein with SHP was reduced by CA feeding, and the protein was identified as psmd1, a component of the 26S proteasomal complex (Fig. 1A; Deng et al. 2007). These results suggested that SHP may be a target for proteasomal degradation and that CA feeding may inhibit the degradation.
Figure 1.
SHP is rapidly degraded by the ubiquitin–proteasome pathway. (A) Experimental outlines for in vivo experiments are shown in Supplemental Figure S1. Briefly, Ad-Flag-SHP or control Ad-empty (E) was injected via the tail vein and, 5 d later, mice were fed normal (−) or CA-supplemented (+) chow for 5 h and liver extracts were prepared. Flag-SHP and associated hepatic proteins affinity-purified on M2 agarose were visualized by silver staining. One band (arrow) was identified as psmd1 by mass spectrometry. (B) HepG2 cells transfected with a Flag-SHP expression plasmid were treated with proteasome inhibitors as indicated for 6 h and subjected to Western analyses. Duplicates are shown. (C) Hepa1c cells were transfected with pcDNA3-Flag-SHP with (+) or without (−) an HA-ubiquitin plasmid and then were treated with vehicle or MG132 for 3 h. Flag-SHP was immunoprecipitated from cell extracts with M2 antibody (IP) and ubiquitinated SHP in the immunoprecipitates was detected by Western analysis (WB). Heavy and light chains of IgG are indicated by asterisks. Positions of ubiquitinated SHP proteins are indicated by a dotted line. Flag-SHP levels in the input are shown in the bottom panel. (D) HepG2 cells infected with Ad-Flag-SHP were metabolically labeled for 30 min followed by chase for the indicated times. Flag-SHP was immunoprecipitated with M2 antibody and detected by autoradiography. (NS) Nonspecific band. Band intensities were measured by densitometry and the intensities relative to the 0-min time point were plotted. (E) HepG2 cells infected with Ad-Flag-SHP were treated with CHX (10 μg/mL) for the indicated times and Flag-SHP levels were detected by Western analyses. Band intensities were quantified as in D. SEM, n = 4. (F) HepG2 cells were treated with CHX for the indicated times and endogenous SHP was immunoprecipitated with SHP antibody or IgG control and detected by Western analyses. As a control, tubulin levels were also detected.
To determine if SHP is a target of proteasomal degradation, we tested the effects of proteasome inhibitors on levels of Flag-SHP expressed in HepG2 cells. Because Flag-SHP expression was controlled by a constitutive CMV promoter, changes in the Flag-SHP levels should result from changes in degradation rather than biosynthesis. Flag-SHP protein levels were robustly increased by MG132, ALLN, or lactacystin treatment (Fig. 1B), and similar results were observed in PHH (Supplemental Fig. S2). These results indicate that SHP is a target of 26S proteasomal degradation in hepatocytes.
Since the 26S proteasome recognizes and degrades proteins that are conjugated with polyubiquitin chains, we next examined whether SHP is ubiquitinated in cells. Because of poor transfection efficiency in HepG2 cells, cell ubiquitination studies were carried out in hepa1c cells. A ladder of high-molecular-weight forms of ubiquitinated SHP proteins was detected in cells transfected with an expression plasmid for HA-ubiquitin, and MG132 treatment increased the ubiquitinated SHP levels as expected (Fig. 1C). These results further support the conclusion that SHP is a target of ubiquitin–proteasomal degradation.
SHP is rapidly degraded with a half-life ∼20–30 min
To determine the rate of degradation of SHP, HepG2 cells expressing Flag-SHP were pulse-labeled with 35S-methionine and then chased by incubation with unlabeled methionine. Flag-SHP levels were decreased by 80% after 1 h of chase and were nearly undetectable after 3 h (Fig. 1D). We also measured the degradation rate by monitoring the decrease of SHP after protein synthesis was blocked by cycloheximide (CHX). Similar to pulse-chase experiments, the half-life of Flag-SHP was ∼30 min after CHX treatment (Fig. 1E).
Since exogenously expressed Flag-SHP might be degraded differently than endogenous SHP, we measured the decrease in endogenous SHP after CHX treatment in HepG2 cells. The half-life of endogenous SHP was ∼20–30 min, similar to that determined for Flag-SHP, whereas levels of tubulin were not changed (Fig. 1F). These results demonstrate that endogenous SHP as well as Flag-SHP is rapidly degraded.
CDCA treatment increases the stability of SHP by blocking ubiquitination
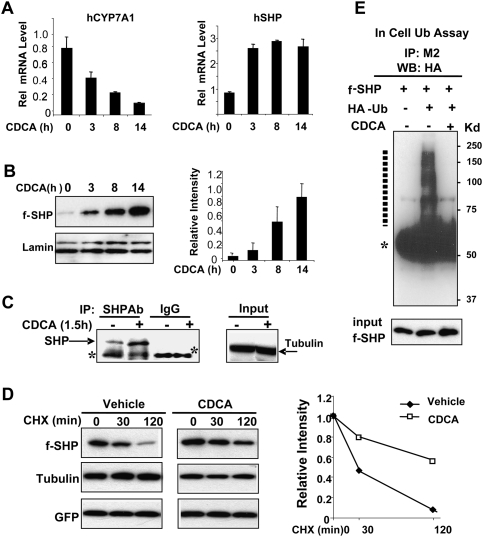
Since bile acids are known to increase SHP levels and resulted in decreased association of psmd1 with SHP in mouse liver (Fig. 1A), we examined the effects of a primary bile acid, chenodeoxy CA (CDCA), on SHP stability. CYP7A1 mRNA levels decreased and SHP mRNA levels were increased after CDCA treatment, as expected (Fig. 2A; Goodwin et al. 2000; Lu et al. 2000). Interestingly, levels of Flag-SHP expressed from the CMV promoter were increased in a time-dependent manner by >10-fold after 14 h of CDCA treatment (Fig. 2B). Notably, while SHP mRNA levels increased less than threefold, increased stability resulted in a >10-fold increase in protein levels (Fig. 2A,B). Increased endogenous SHP levels were also detected after CDCA treatment in HepG2 cells (Fig. 2C).
Figure 2.
CDCA treatment increases SHP stability by inhibiting ubiquitination. (A) HepG2 cells were treated with 50 μM CDCA for the indicated times and mRNA levels of CYP7A1 and SHP were measured by qRT–PCR and normalized to those of 36B4. SEM, n = 3. (B) HepG2 cells expressing Flag-SHP were treated with CDCA for the indicated times and subjected to Western analysis. The average intensity for Flag-SHP relative to the corresponding intensity for lamin was quantified. SEM, n = 4. (C) HepG2 cells were treated with vehicle or CDCA and endogenous SHP levels were detected by immunoprecipitation followed by Western analyses. The control tubulin levels were detected from the input. (D) HepG2 cells infected with Ad-Flag-SHP were treated with CHX for the indicated times in the presence of CDCA (+) or vehicle (−) and protein levels were detected by Western analyses. Band intensities were measured by densitometry and the intensities relative to the 0-min time point were plotted. Consistent results were observed from two independent CHX experiments. (E) Hepa1c cells transfected with pcDNA3-Flag-SHP with (+) or without (−) a ubiquitin plasmid were treated with vehicle or CDCA for 3 h. In-cell ubiquitination assays were done as described in the legend for Figure 1. Ubiquitinated Flag-SHP forms are indicated by a dotted line, and heavy chain of IgG is indicated by an asterisk.
To examine the effect of CDCA treatment on the rate of degradation of SHP, we examined SHP protein levels after CHX treatment. CDCA treatment reduced the degradation rate so that ∼50% of Flag-SHP was still detected at 120 min after CHX treatment, while in cells not treated with CDCA, the half-life was ∼30 min (Fig. 2D). Levels of tubulin as a loading control and GFP as a control for adenoviral infection efficiency were not affected by CHX treatment (Fig. 2D).
If CDCA reduces proteasomal degradation of SHP, it would be expected that ubiquitination should also be reduced. CDCA treatment abolished ubiquitinated forms of Flag-SHP present in HepG2 cells (Fig. 2E). These results suggest that SHP is rapidly degraded by proteasomal degradation and that CDCA treatment increases SHP stability by inhibiting ubiquitination and subsequent proteasomal degradation.
The ERK pathway is critical for SHP stability
Since regulation of ubiquitin-mediated proteasomal degradation by phosphorylation is well known (Pickart 2001; Brooks and Gu 2003; Sundqvist et al. 2005) and bile acids activate kinase signaling pathways such as JNK and ERK Ser/Thr kinases (Gupta et al. 2001; Wang et al. 2002; Holt et al. 2003; Dent et al. 2005), we examined whether phosphorylation is important for SHP stability. Treatment with calyculin A, a Ser/Thr-specific phosphatase inhibitor, substantially increased Flag-SHP protein levels in CDCA-treated cells (Supplemental Fig. S3), suggesting that phosphorylation of SHP is involved in its stability.
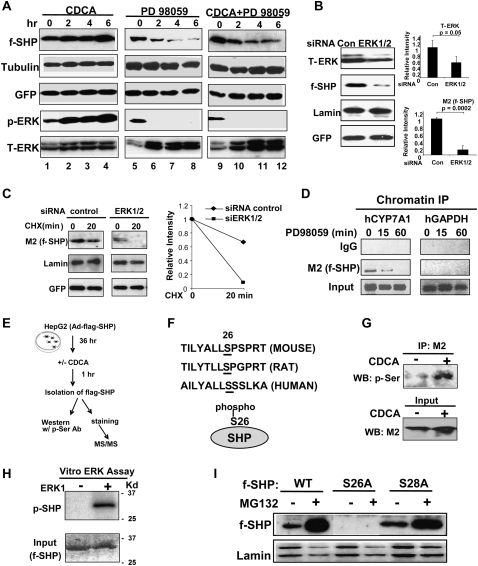
To determine whether known bile acid-regulated kinase signaling pathways are involved in stabilization of SHP, HepG2 cells were treated with inhibitors of JNK (SP600125), ERK (PD98059), and p38 kinase (SB203580). JNK has been shown to be prominently involved in bile acid and FGF19-mediated CYP7A1 repression (Holt et al. 2003; Yu et al. 2005). Surprisingly, inhibition of JNK or p38 kinase did not reduce Flag-SHP levels, but instead increased Flag-SHP levels in cells treated with either vehicle or CDCA (Supplemental Fig. S4). In a sharp contrast, treatment with an ERK inhibitor, PD98059, resulted in a time-dependent decrease in Flag-SHP levels in cells treated with either CDCA or vehicle (Fig. 3A, lanes 5–12). Furthermore, phospho-ERK levels were decreased to undetectable levels by 2 h, whereas total ERK levels were not reduced (Fig. 3A, lanes 5–12). The levels of the control proteins, tubulin or GFP, were not changed. CDCA treatment of HepG2 cells resulted in an increase in phospho-ERK levels in parallel with increased Flag-SHP levels (Fig. 3A).
Figure 3.
ERK pathway is critical for SHP stability and Ser26 is the ERK target. (A) HepG2 cells infected with Ad-Flag-SHP were treated as indicated (50 μM CDCA, 10 μM PD98059) for 0, 2, 4, and 6 h and subjected to Western analysis. (B,C) HepG2 cells were transfected with siRNA (30 ng) for ERK1/2 or control siRNA and, 24 h later, cells were infected with Ad-Flag-SHP, and then, 24 h later, cells were subjected to either Western analysis (B) or CHX experiments (C). Band intensities were determined using ImageJ and the values for control samples were set to 1. Statistical significance was determined by the Student's t-test (SEM, n = 3). (D) HepG2 cells were infected with Ad-Flag-SHP and then subjected to ChIP assays using M2 antibody. Associations of Flag-SHP with the CYP7A1 promoter (−320 to +10) and control GAPDH coding region (+181 to +459) are shown. (E) Experimental outlines. (F) A predicted ERK site in mouse, rat, and human SHP is shown. Ser26 identified as the major ERK site by MS/MS is underlined. (G) Phosphorylated Ser levels in SHP were detected by Western blotting using phospho-Ser antibody. Flag-SHP levels in input are shown in the bottom panel. (H) In vitro ERK assay: Flag-SHP was expressed in HepG2 cells by adenoviral infection and purified by M2 agarose and incubated with γ32P-ATP with (+) or without (−) purified ERK. Phosphorylated SHP was detected by autoradiography and total SHP was detected by colloidal staining. (I) HepG2 cells were transfected with expression plasmids for Flag-SHP wild type, S26A, or S28A and then treated with 5 μM MG132 for 6 h, and cell extracts were subjected to Western analysis.
Similar to the results from HepG2 cells, Flag-SHP and active phospho-ERK levels were increased after CDCA treatment in PHH (Supplemental Fig. S5). In control experiments, SHP mRNA levels were increased, whereas CYP7A1 levels were decreased, in PHH after CDCA treatment (Supplemental Fig. S6). These results indicate that CDCA treatment increases the levels of activated phospho-ERK in hepatocytes and further suggest that CDCA-mediated activation of ERK results in increased SHP stability.
To directly demonstrate that the ERK pathway is critical in regulating SHP stability, endogenous ERK1/2 in HepG2 cells were down-regulated by siRNA and Flag-SHP protein levels were measured. ERK1/2 levels were significantly reduced in cells overexpressing siRNA for ERK and down-regulation of endogenous ERK1/2 dramatically decreased Flag-SHP protein levels (Fig. 3B). In CHX experiments, down-regulation of ERK1/2 increased the degradation rate so that Flag-SHP was not detectable at 20 min after CHX treatment (Fig. 3C). These results demonstrate that the ERK pathway is critical for SHP stability.
To further examine whether ERK is critical for SHP protein function, the association of SHP with the promoter of CYP7A1, a well-known SHP target gene, was monitored in HepG2 cells after treatment with PD98059, an ERK inhibitor. Association of Flag-SHP with the promoter was nearly abolished 1 h after PD98059 treatment (Fig. 3D), consistent with the decreased levels of SHP in these cells. These results demonstrate that the ERK-mediated decrease in SHP levels is functionally significant.
Ser26 in SHP is the ERK phosphorylation site
To identify phosphorylated amino acid residues, HepG2 cells were infected with Ad-Flag-SHP and treated with vehicle or CDCA, and Flag-SHP was isolated (Fig. 3E; Supplemental Fig. S7). MS/MS analysis revealed that Ser26 was phosphorylated after CDCA treatment (Fig. 3F; Supplemental Fig. S7). Ser26 is within the sequences 25-LSPSP-29, which is a predicted ERK site, Pro-neighboring Ser/Thr, in SHP. Consistent with these results, Western analysis showed that CDCA treatment increased phospho-Ser levels in SHP (Fig. 3G). To directly determine whether SHP can be phosphorylated by ERK, in vitro kinase assays were performed. Phosphorylated SHP was detected after incubation of Flag-SHP with ERK and γ32P-ATP by autoradiography (Fig. 3H) or by Western analysis with phospho-Ser antibody (Supplemental Fig. S8).
We further examined the role of Ser26 in SHP stability by mutational analysis. While mutation of Ser28 to alanine did not change SHP stability, as almost comparable with wild-type SHP, mutation of Ser26 to either alanine or glutamate (data not shown) resulted in undetectable levels of Flag-SHP (Fig. 3I). These results demonstrate that Ser26 is phosphorylated in response to bile acid signaling and further suggest that Ser26 is critical for SHP stability.
Bile acid-induced FGF19 increases SHP stability through ERK
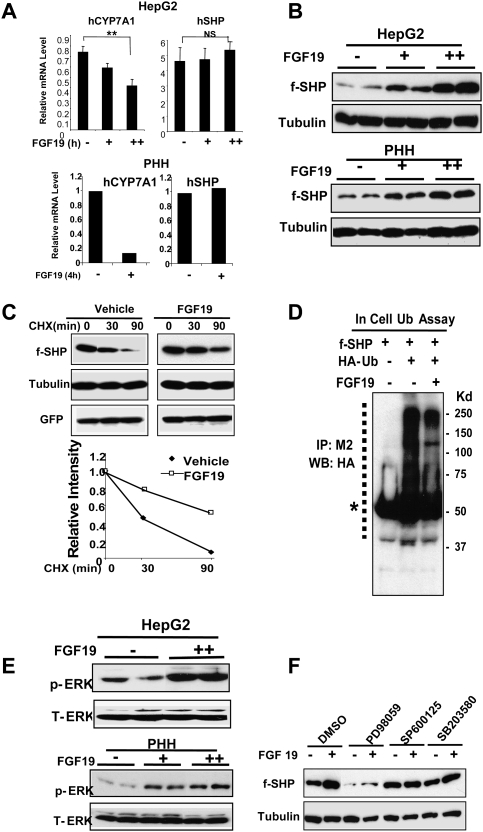
Bile acid-activated FXR induces the expression of FGF15/19 in the small intestine, and the secreted FGF15/19 acts at the liver to inhibit CYP7A1 transcription (Inagaki et al. 2005). This inhibition was substantially reversed in SHP-null mice, but surprisingly, the SHP mRNA levels in the liver of wild-type mice were not increased by FGF15/19 treatment. One possibility is that FGF15/19 increases SHP stability rather than gene induction. Consistent with previous studies (Inagaki et al. 2005), CYP7A1 mRNA levels were significantly decreased after FGF19 treatment, whereas SHP mRNA levels were not significantly changed in HepG2 cells or in PHH by treatment with FGF19 (Fig. 4A). In contrast, treatment with FGF19 substantially increased Flag-SHP protein levels in both HepG2 and PHH (Fig. 4B), suggesting that the major effect of FGF19 is on the stability of SHP.
Figure 4.
FGF19 increases SHP stability by inhibiting ubiquitination of SHP and activating ERK. (A) HepG2 cells or PHH were treated with purified recombinant FGF19 (0, 40 ng/mL [+], 100 ng/mL [++]) for 3 h, and the mRNA levels of CYP7A1 and SHP were detected by qRT–PCR and normalized to those of 36B4. SEM, n = 3. (B) HepG2 cells or PHH were infected with Ad-Flag-SHP and then treated with FGF19 for 3 h and subjected to Western analyses. Duplicates are shown. (C) HepG2 cells infected with Ad-Flag-SHP were treated with CHX for the indicated times in the presence of FGF19 (+) or vehicle (−) and subjected to Western analyses. Band intensities relative to the 0-min time point were plotted. (D) Hepa1c cells were transfected with expression plasmids as indicated and, 36 h later, cells were treated with FGF19 for 30 min and subjected to in-cell ubiquitination assays as described in the legend for Figure 1. (E) HepG2 cells were treated with FGF19 for 3 h and phosphorylated ERK (p-ERK) and total ERK (T-ERK) levels were detected by Western analyses. Duplicates are shown. (F) HepG2 cells infected with Ad-Flag-SHP were pretreated with the indicated kinase inhibitors for 30 min and further treated with FGF19 (+) or vehicle PBS (−) for 3 h and then subjected to Western analysis.
The half-life of SHP was markedly increased after FGF19 treatment in HepG2 cells treated with CHX (Fig. 4C). Consistent with the decreased degradation, ubiquitinated SHP levels were substantially reduced in cells after FGF19 treatment (Fig. 4D), suggesting that FGF19 increases SHP stability by inhibiting ubiquitination and subsequent proteasomal degradation.
As observed with CDCA treatment, FGF19 treatment increased the levels of phosphorylated ERK in HepG2 cells or PHH, whereas total ERK levels were not changed (Fig. 4E). Inhibition of ERK, but not JNK and p38, resulted in dramatically decreased levels of SHP in both untreated and FGF19-treated cells (Fig. 4F), as was observed after CDCA treatment (Fig. 3B). In control experiments, phosphorylated ERK levels were undetectable after treatment with ERK inhibitor in FGF19-treated HepG2 cells (Supplemental Fig. S9). These results strongly suggest that FGF19 increases SHP stability by inhibiting ubiquitination and proteasomal degradation, possibly as a result of ERK activation.
Lysine residues 122 and 123 in SHP are major ubiquitination sites
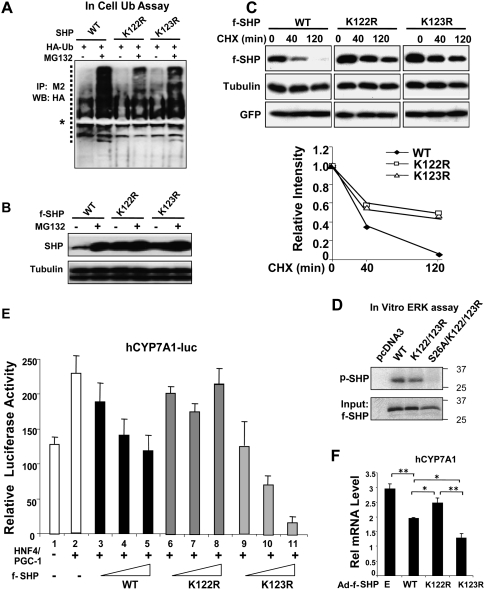
To identify lysine residue targets for ubiquitination, each of the six lysine residues present in SHP were substituted with arginines and in-cell ubiquitination assays were performed. Ubiquitinated SHP levels were reduced by mutation of K122 or K123 (Fig. 5A), whereas they were not markedly reduced in mutation of other lysines (data not shown). The protein levels of these mutants were elevated in cells before treatment with MG132 compared with wild-type SHP and were only modestly increased after CDCA treatment (Fig. 5B). Consistent with the reduced ubiquitination, the half-lives of the K122 and K123 mutants were substantially increased compared with that of wild type (Fig. 5C).
Figure 5.
Lys122 and Lys123 are major ubiquitination sites in SHP. (A) Hepa1c cells transfected with expression vectors as indicated were treated with vehicle or 5 μM MG132 for 6 h and subjected to in-cell ubiquitination assays as described in the legend for Figure 1. (B) HepG2 cells were transfected with expression vectors as indicated and treated with 5 μM MG132 (+) or vehicle (−) for 6 h and then subjected to Western analysis. (C) HepG2 cells infected with Ad-Flag-SHP wild type (WT) or mutants were treated with CHX for the indicated times and subjected to Western analyses. Band intensities relative to the 0-min time point were plotted. (D) Wild-type and indicated Flag-SHP mutant proteins were synthesized using in vitro transcription and translation system. Flag-SHP was immunoprecipitated by M2 agarose and subjected to in vitro ERK kinase assay as described in the legend for Figure 3. Phosphorylated SHP was detected by autoradiography and Flag-SHP levels in the input samples were detected by Western analysis. The pcDNA3 serves a negative control. (E) HepG2 cells were cotransfected with 250 ng of CYP7A1-luc reporter and expression vectors for CMV β-galactosidase as an internal control, 100 ng of HNF-4 and PGC-1α, as well as 25–200 ng of pcDNA3-Flag-SHP wild type or mutants, and then cells were harvested for reporter assays. The triangles represent increasing amounts of the Flag-SHP vectors. The values for firefly luciferase activities were normalized by dividing by the β-galactosidase activities. SEM, n = 3. (F) HepG2 cells were infected with Ad-virus and, 36 h later, cells were further subjected to qRT–PCR. Statistical significance was measured using the Student's t-test. (*) P < 0.05; (**) P < 0.01; SEM, n = 4.
Mutation of the major ERK phosphorylation site Ser26 dramatically destabilized SHP (Fig. 3I). However, additional mutations at both ubiquitination sites, K122 and K123, substantially reversed the destabilization by the S26 mutation (Supplemental Fig. S10). Furthermore, while robust phosphorylation of SHP wild type and the K122/123 SHP mutant was detected, phosphorylation of the S26/K122/K123 mutant was not detected (Fig. 5D). These results, together with MS/MS analyses, demonstrate that K122 and K123 are major ubiquitination sites and Ser26 is a major ERK phosphorylation site in SHP.
Mutation of K122/K123 alters the repression activity of SHP
Since mutation of K122 and K123 to Arg increased SHP protein levels, we tested whether the repression activity of SHP was also increased. Exogenous expression of the K123R SHP mutant enhanced the suppression of the CYP7A1-luc reporter activity (Fig. 5E, cf. lanes 3–5 and 9–11). Surprisingly, suppression was not enhanced by the K122R mutant, and in fact was impaired (Fig. 5E, lanes 6–8) despite increased protein levels (Fig. 5B,C). These data are consistent with the hypothesis that the mutation at K123 increases the stability of SHP without reducing its repression activity, while mutation at K122 increases the stability of SHP, but also inhibits its repression activity. Since K122 is present in the nuclear receptor-interacting region in the central domain of SHP, we tested whether the K122R mutant shows impaired interaction with HNF-4, a well-known SHP-interacting nuclear receptor (Lee et al. 2000; Lee and Moore 2002). In vitro GST pull-down assays showed that the K122R mutant efficiently interacted with HNF-4 (Supplemental Fig. S11), so that impaired interaction with nuclear receptors does not explain the decreased activity of the K122R mutant.
We further examined the effects of ectopic expression of SHP wild type or mutants using adenoviral infection on expression of endogenous CYP7A1 in HepG2 cells. Similar infection efficiencies were obtained as monitored by detection of GFP (Supplemental Fig. S12). Consistent with the results from reporter assays, exogenous expression of Flag-SHP wild type reduced mRNA levels of endogenous CYP7A1, and the K123R mutant significantly enhanced SHP repression of CYP7A1 (Fig. 5F). In contrast, exogenous expression of the K122R mutant decreased CYP7A1 mRNA levels significantly less than wild type, even though K122R levels were increased (Fig. 5F), consistent with the lack of enhanced suppression by this mutant in the reporter assays (Fig. 5E). These results suggest that the K123 mutant results in increased stability of an active protein, while the K122 mutant results in increased stability of SHP with reduced activity.
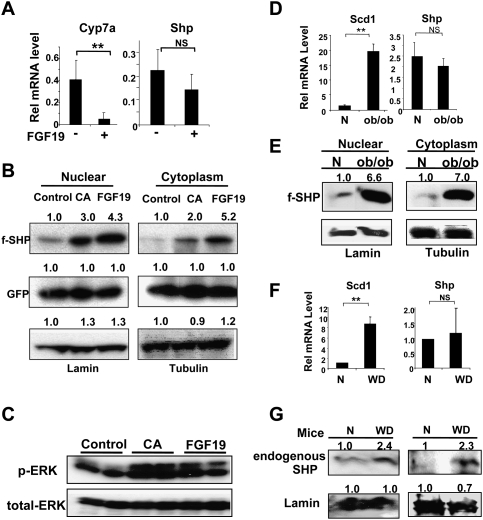
CA feeding and FGF19 treatment increase SHP stability in vivo
To determine the in vivo relevance of the findings from cell culture studies, levels of Flag-SHP in mouse liver after treatment with bile acids or FGF19 were determined. Consistent with previous reports (Goodwin et al. 2000; Lu et al. 2000; Kerr et al. 2002; Wang et al. 2002), Cyp7a1 mRNA levels were decreased, whereas the Shp mRNA levels were increased by about twofold after 14 h of CA feeding (data not shown). In contrast, while Cyp7a1 mRNA levels were decreased by FGF19 treatment, Shp mRNA levels were not increased by FGF19 treatment (Fig. 6A). However, Flag-SHP levels in both nuclear and cytoplasmic extracts were substantially increased fourfold to fivefold after FGF19 treatment (Fig. 6B), suggesting that treatment with bile acids or FGF19 increased SHP stability without markedly changing the cellular location of the protein. These results indicate that in the case of CA feeding, both regulation of SHP stability and gene induction contribute to increased SHP levels. In contrast, for FGF19, effects on SHP stability predominate with little or no effect on SHP gene induction. These results are consistent with results from cultured cell studies and support the conclusion that SHP stability is substantially increased by bile acids or FGF19 treatment.
Figure 6.
Hepatic SHP stability is increased by CA feeding or FGF19 treatment in vivo and is abnormally elevated in metabolic disease model mice. (A) Mice were treated with vehicle PBS (−) or FGF19 (+) via the tail vein and, 6 h later, livers were collected for qRT–PCR. (B) Mice were injected with Ad-Flag-SHP and, 5 d later, fed normal control chow or 0.5% CA-supplemented chow (CA) for 14 h, or treated with FGF19 for 6 h. Nuclear and cytoplasmic extracts were prepared and Flag-SHP and lamin (as a nuclear protein control), tubulin (as a cytoplasmic protein control), and GFP (as a monitor of infection efficiency) were detected by Western analyses. Consistent results were observed from three sets of mice. (C) Mice were fed control chow or CA chow or were injected with FGF19 and livers were collected for Western analysis. (D) Livers from normal wild-type CV57 mice (N) or ob/ob mice were collected for qRT–PCR. (E) Wild-type normal (N) or ob/ob mice were injected with Ad-Flag-SHP and, 5 d later, cytoplasmic and nuclear extracts were prepared for Western blotting. Consistent results were observed from two sets of mice. (F) Livers from normal mice (N) or mice fed a high-fat and high-calorie western-style diet (WD) for 16 wk were collected for qRT–PCR. (G) Diet-induced obese mouse model: Livers from mice fed normal chow (N) or fed a chronic western diet (WD) for 16 wk were collected. Endogenous Shp levels were detected by Western analyses using SHP antibody. Results from two sets of mice are shown. (B,E,G) Band intensities were determined using ImageJ and the values for control or normal samples were set to 1. (A,D,F) Statistical significance was determined by the Student's t-test (SEM, n = 3). (**) P < 0.01; (NS) statistically not significant.
Since active phosphorylated ERK levels were increased in HepG2 and PHH and inhibition of ERK dramatically destabilized SHP (Fig. 3), we examined if activated ERK levels are also increased in mouse liver after CA feeding or FGF19 treatment. While total ERK levels were similar, the phosphorylated active form of ERK in liver was substantially increased after CA feeding or FGF19 treatment (Fig. 6C), suggesting that ERK is activated in both cases. These results are consistent with results from cultured cell studies and support the conclusion that SHP stability is substantially increased by bile acids or FGF19 treatment through activation of ERK.
SHP stability is elevated in ob/ob mice and diet-induced obese mice
Reduced lipid levels and increased insulin sensitivity were observed in SHP-null mice (Wang et al. 2005) and, conversely, elevated hepatic lipid levels were observed in transgenic mice overexpressing Flag-SHP (Boulias et al. 2005). These transgene studies suggest that highly elevated SHP levels could underlie abnormal hepatic metabolism in metabolic disease states. To test this possibility, we examined the mRNA and protein levels of SHP in ob/ob mice and mice fed a chronic western-style diet, which are animal models for fatty liver (liver steatosis) and type II diabetes. In ob/ob mice, the mRNA levels of a hepatic lipogenic marker, stearoyl coA desatrurase1 (Scd1), were dramatically elevated, as expected (Fig. 6D; Huang et al. 2007). In contrast, Shp mRNA levels were not significantly changed in both ob/ob mice (Fig. 6D) and in mice fed a chronic western diet (Fig. 6F). Interestingly, Shp mRNA levels in Balb/C mouse livers were markedly decreased compared with those in CV57 mice and ob/ob mice (Supplemental Fig. S13), emphasizing the metabolic differences among strains and the need to use congenic mice as controls. Surprisingly, in contrast to the unchanged mRNA levels, Flag-SHP levels were dramatically elevated in both nuclear and cytoplasmic hepatic extracts from ob/ob mice compared with congenic CV57 mice (Fig. 6E). Also, in mice fed a chronic western-style diet, endogenous SHP protein levels were increased by more than twofold (Fig. 6G), even though the mRNA levels were not increased (Fig. 6F). The mechanism underlying elevated SHP stability in these obese mice is not clear, but these intriguing results provide a potential link between abnormally elevated SHP stability and metabolic disease states.
Discussion
Negative feedback regulation of bile acid biosynthesis and its importance in maintaining cholesterol and bile acid homeostasis has been known for many years. A major advance in understanding the mechanism of this regulation was the demonstration that the nuclear bile acid receptor FXR induces the expression of an orphan nuclear receptor, SHP, which in turn represses the expression of CYP7A1, the first and rate-limiting enzyme in the bile acid synthetic pathway (Goodwin et al. 2000; Lu et al. 2000). It was also shown that cell signaling mechanisms other than this nuclear receptor cascade modulate SHP regulation of CYP7A1 (Gupta et al. 2001; Kerr et al. 2002; Wang et al. 2002; Holt et al. 2003), which raises the possibility that post-translational modifications might regulate SHP function. Here, we report that, in addition to effects on gene induction by the FXR, hepatic bile acids also dramatically stabilize SHP protein to suppress hepatic bile acid biosynthesis. Furthermore, bile acid-induced intestinal FGF19 acts at the liver by increasing hepatic SHP stability, without increasing SHP gene induction.
As summarized in Figure 7, in the basal state, SHP is degraded rapidly in hepatocytes by the ubiquitin–proteasomal pathway, with a half-life of <30 min. This short half-life would be consistent with SHP's role as a critical metabolic regulator whose activity must be rapidly and tightly modulated in response to hormonal, nutritional, and other cellular signals. When hepatic bile acid levels are elevated, bile acids increase SHP protein levels by blocking ubiquitination and proteasomal degradation. CDCA treatment resulted in increased levels of phosphorylated ERK, inhibition of ERK resulted in increased degradation of SHP, and mutation of Ser26, which is phosphorylated by ERK, resulted in a dramatic increase in SHP degradation, all of which are consistent with the hypothesis that the effects of CDCA on SHP stability are mediated by activation of ERK. These observations are consistent with the demonstration that ubiquitination in many systems is dependent on upstream phosphorylation-dependent processes (Pickart 2001; Brooks and Gu 2003; Sundqvist et al. 2005). These studies demonstrate for the first time that increased stability of SHP plays an important role in increased SHP action in the liver in response to elevated bile acids, in addition to the increased SHP gene expression.
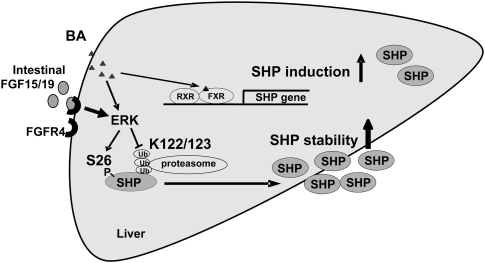
Figure 7.
Bile acids and FGF19 increase stability of hepatic SHP by inhibiting ubiquitin–proteasomal degradation and activating ERK pathway. SHP is rapidly degraded by the ubiquitin–proteasomal pathway with a half-life <30 min. Bile acids not only increase SHP gene induction by activation of the nuclear bile acid receptor FXR, but also substantially increase SHP stability by activating ERK and inhibiting proteasomal degradation. Treatment with a primary bile acid, CDCA, increases phosphorylation of SHP at Ser26 and inhibits ubiquitination at Lys122/Lys123, which results in increased SHP stability. Bile acid-induced intestinal FGF19 predominantly increases SHP stability, with little effect on SHP gene induction, by activating ERK and inhibiting ubiquitination and proteasomal degradation.
A second major advance in understanding the mechanism of feedback inhibition of bile acid biosynthesis was the observation that bile acid-activated FXR induces the expression of FGF15/19 in the intestine (Inagaki et al. 2005). The secreted FGF19 acts at the liver to suppress CYP7A1 expression by a mechanism that appears to be SHP-dependent, since the suppression is decreased in SHP-null mice. It was surprising, therefore, that the hepatic levels of SHP mRNA were not increased by treatment with FGF19, so the reason for SHP dependence was somewhat puzzling. Our present studies provide an explanation. FGF19 dramatically stabilizes SHP in the liver, resulting in increased SHP levels even though SHP mRNAs are not increased, which explains the SHP dependence. The mechanism of FGF19 stabilization of SHP is very similar to that of bile acids. In both cases, ubiquitination is decreased and ERK is activated, so a plausible model is that FGF19 increases ERK-mediated phosphorylation of SHP, which results in decreased ubiquitination and proteasomal degradation. The difference between bile acids and FGF19 is that bile acids increase both SHP gene expression and SHP stability, while FGF19 acts only through effects on SHP stability.
These studies have further established that increased SHP stability is associated with conditions of abnormal metabolic states in mice. An important role for SHP in lipid and glucose metabolism was established by the reduced liver and serum lipid levels and increased insulin sensitivity observed in SHP-null mice (Kerr et al. 2002; Wang et al. 2005, 2006) and, conversely, elevated hepatic lipid levels observed in transgenic mice overexpressing Flag-SHP (Boulias et al. 2005)). In ob/ob mice, a mouse model of fatty liver disease and type II diabetes, high lipid and glucose levels in both liver and serum were reduced by crossing the mice with SHP-null mice, suggesting that the abnormal lipid and glucose levels were dependent on SHP (Huang et al. 2007). We showed in two metabolic disease models, ob/ob mice and diet-induced obese mice, that SHP levels are increased compared with control mice—a result of increased stability of SHP. In both cases, SHP mRNA levels were not increased so that the major defect in regulation of SHP is in the stability of the protein. The mechanism of the increased stability of SHP is not clear in these disease models, but SHP stability is clearly an important component underlying the metabolic dysfunction.
The major sites of ubiquitination in SHP are K122 and K123. Mutation of these lysine residues to arginine is expected to increase basal levels of SHP, if these sites are ubiquitinated. Bile acid treatment results in only modest additional increases in the levels of the mutants if bile acids act by inhibiting ubiquitination at these sites. In functional assays, the repression activity of the K123R mutant was enhanced compared with wild type, consistent with elevated levels of this mutant. Unexpectedly, however, the repression activity of the K122R mutant was decreased even though levels of protein were elevated. The explanation for this observation is unclear; in vitro GST pull-down assays suggest that decreased repression activity of the K122R mutant does not result from impaired interaction with nuclear receptors. Post-translational modification of K122 to a modification other than ubiquitination itself might be important for SHP inhibitory activity. For example, K122 could be a target of sumoylation, which might be essential for SHP repression activity, as has been shown in other transcriptional activators like PPARγ (Pascual et al. 2005).
SHP is emerging as a critical regulator for multiple metabolic pathways, including cholesterol/bile acid, fatty acid/triglyceride, and glucose metabolism (Bavner et al. 2005; Boulias et al. 2005; Wang et al. 2005). Dysregulation of these metabolic pathways underlies major metabolic diseases in humans, such as liver steatosis, obesity, diabetes, and cardiovascular disease. Our studies show that regulation of SHP stability is important both in physiological regulation of metabolic homeostasis and in pathological conditions. Components of the pathways involved in the regulation of SHP stability, therefore, may be attractive targets for the development of therapeutic agents that modulate SHP stability to treat these metabolic disorders.
Materials and methods
Cell culture and reagents
PHH were obtained from the Liver Tissue Procurement and Distribution System of the National Institutes of Health (S. Strom, University of Pittsburgh, PA). HepG2 cells (American Type Culture Collection [ATCC] HB8065) and mouse Hepa1c1c7 cells (ATCC CRL2026) were maintained as described (Kemper et al. 2004; Fang et al. 2007). CDCA, CHX, proteasome inhibitors, and M2 antibody were purchased from Sigma, Inc., and kinase inhibitors were purchased from Calbiochem, Inc. ERK siRNA and control siRNA were purchased from Applied Biosystems. Antibodies for SHP, ERK, HA, GFP, tubulin, and lamin were purchased from Santa Cruz Biotechnologies.
Plasmid and adenoviral vector constructs
For constructing the pcDNA3 Flag-mouse SHP, pcDNA3 Flag-SIRT1 plasmid, which was provided by Dr. W. Gu (Columbia University), was digested with EcoRI and NotI to remove the SIRT1 sequence, and then a 0.9-kb fragment containing SHP cDNA was inserted into the resulting Flag-pcDNA3 vector. Flag-SHP mutants were constructed by site-directed mutagenesis (Stratagene, Inc.) and confirmed by sequencing. Ad-Flag-SHP wild type and mutants were constructed with the Ad-CMV-Track system and the adenovirus vectors, which also express GFP.
Animal experiments
For adenoviral experiments, mice were injected via the tail vein with ∼0.5 × 109 active viral particles in 200 μL of PBS and, 4–7 d after infection, the mice were fed normal chow or chow supplemented with 0.5% CA (Harland Teklad TD05271) for 3–24 h For western diet mouse experiments, mice were fed normal chow or a high-fat chow (42% from fat; Harland Teklad TD88137) for 16 wk. All the animal use and adenoviral protocols were approved by the Institutional Animal Care and Use and Institutional Biosafety Committees at University of Illinois at Urbana-Champaign and were in accordance with National Institutes of Health guidelines.
Hepatic Flag-SHP complex purification and mass spectrometry analysis
Flag-SHP was expressed in mouse liver by adenoviral infusion and isolated by binding to M2 agarose. Briefly, 10 mg of proteins of liver extracts were incubated with M2 agarose in binding buffer (20 mM Tris-HCl at pH 7.4, 1 mM EDTA, 0.1% NP40, 15% glycerol, plus 0.1% SDS in the phosphorylation study) for 4 h and the M2 agarose was washed with binding buffer six times. Proteins associated with Flag-SHP were separated by SDS-PAGE and detected by silver staining. Protein bands were excised and subjected to MS/MS analysis.
Pulse-chase and CHX experiments
HepG2 cells were infected with Ad-Flag-SHP and were incubated in methionine-free medium for 2 h. Trans-label mixture (BP Biomedical, Inc.) containing 35S-methionine was added for 30 min and then cells were cultured in medium containing 10× unlabeled methionine for up to 3 h. Flag-SHP was immunoprecipitated with M2 antibody in RIPA buffer (50 mM Tris at pH 7.5, 1 mM EDTA, 1% NP40, 1% sodium deoxycholate, 0.1% SDS) and radioactive Flag-SHP was detected by autoradiography. For endogenous experiments, HepG2 cells in 10-cm plates (per group) were treated with CHX (10 μg/mL), SHP was immunoprecipitated with SHP antibody (sc-30169) and washed in RIPA buffer, and SHP was detected using SHP antibody (sc-30169 or sc-15283).
In-cell ubiquitination assays
Because of poor transfection efficiency in HepG2 cells, cell ubiquitination studies were carried out in hepa1c1c7 cells. Cells were transfected with expression plasmids for Flag-SHP and HA-ubiquitin and, 24–36 h later, Flag-SHP was immunoprecipitated with M2 antibody in RIPA buffer. Samples were subjected to SDS-PAGE and HA-ubiquitinated Flag-SHP was detected by Western analyses using HA antibody.
MS/MS analysis and in vitro ERK assay
Flag-SHP was expressed in HepG2 cells by adenoviral infection and, 36 h later, cells were treated with either vehicle or 50 μM CDCA together with 5 μM MG132 to block proteasomal degradation. Flag-SHP was purified by binding to M2 agarose to near 90% homogeneity and subjected to MS/MS analysis (Supplemental Fig. S6). For in vitro ERK assays, Flag-SHP that had been bound to M2 agarose was incubated with 10 μCi γ32P-ATP with 20 ng of purified ERK (Millipore, Inc.) in 25 mM Tris-HCl (pH 7.5), and 20 μM EGTA for 10 min at 30°C. After Flag-SHP attached to the M2 agarose was extensively washed, radioactive Flag-SHP was detected by autoradiography. Alternatively, Flag-SHP was incubated with unlabeled ATP and ERK, and phosphorylated proteins were detected by Western analysis using phospho-Ser-antibody (Millipore, Inc.).
Chromatin immunoprecipitation (ChIP) assay
HepG2 cells were infected with Ad-Flag-SHP and, 36 h later, cells were treated with 10 μM PD98059 for 0, 15, and 60 min and cells were subjected to ChIP assays as described previously (Kemper et al. 2004; Miao et al. 2006; Fang et al. 2007; Ponugoti et al. 2007)
Real time quantitative RT–PCR (qRT–PCR) analysis
Total RNA was isolated using Trizol reagent, cDNA was synthesized using a reverse transcriptase kit (Promega, Inc.), and qRT–PCR was performed with an iCycler iQ (Bio-Rad, Inc.). The amount of mRNA for each gene was normalized to that of 36B4 mRNA.
Acknowledgments
We thank Drs. W. Gu, Z. Ronai, and R. Hey for kindly providing plasmids for this study. We thank the Liver Tissue Procurement and Distribution System of the National Institutes of Health (University of Pittsburgh, PA) for providing human primary hepatocytes. Special thanks to B. Kemper for critical comments on the manuscript. This study was supported by grants DK071662 to H.E.X. and NIH DK062777 and AHA 0756028Z to J.K.K. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 to T.D.V. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1773909.
Supplemental material is available at http://www.genesdev.org.
References
- Bavner A., Sanyal S., Gustafsson J.A., Treuter E. Transcriptional corepression by SHP: Molecular mechanisms and physiological consequences. Trends Endocrinol. Metab. 2005;16:478–488. doi: 10.1016/j.tem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Boulias K., Katrakili N., Bamberg K., Underhill P., Greenfield A., Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Chiang J.Y.L. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr. Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- Deng S., Zhou H., Xiong R., Lu Y., Yan D., Xing T., Dong L., Tang E., Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res. Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- Dent P., Fang Y., Gupta S., Studer E., Mitchell C., Spiegel S., Hylemon P.B. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 2005;42:1291–1299. doi: 10.1002/hep.20942. [DOI] [PubMed] [Google Scholar]
- Fang S., Miao J., Xiang L., Ponugoti B., Treuter E., Kemper J.K. Coordinated recruitment of histone methyltransferase G9a and other chromatin modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell. Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B., Jones S.A., Price R.R., Watson M.A., McKee D.D., Moore L.B., Galardi C., Wilson J.G., Lewis M.C., Roth M.E., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stravitz T., Dent P., Hylemon P. Down-regulation of cholesterol 7 a-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-jun N-terminal kinase pathway. J. Biol. Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- Holt J.A., Luo G., Billin A.N., Bisi J., McNeill Y.Y., Kozarsky K.F., Donahee M., Wang D.Y., Mansfield T.A., Kliewer S.A., et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes & Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten S.M., Watanabe M., Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Iqbal J., Saha P.K., Liu J., Chan L., Hussain M.M., Moore D.D., Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- Inagaki T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G., Luo G., Jones S.A., Goodwin B., Richardson J.A., et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Jelinek D.F., Andersson S., Slaughter C.A., Russell D.W. Cloning and regulation of cholesterol 7 α-hydroxylase, the rate-limiting enzyme in bile acid biosynthesis. J. Biol. Chem. 1990;265:8190–8197. [PMC free article] [PubMed] [Google Scholar]
- Kemper J., Kim H., Miao J., Bhalla S., Bae Y. Role of a mSin3A–Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell. Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr T.A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D.W., Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Moore D.D. Dual mechanism for repression of the monomeric orphan receptor liver receptor homologous protein-1 (LRH-1) by the orphan small heterodimer partner (SHP) J. Biol. Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- Lee Y., Dell H., Dowhan D.H., Hadzopoulou-Cladaras M., Moore D.D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: Two mechanisms for repression. Mol. Cell. Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Miao J., Fang S., Bae Y., Kemper J.K. Functional inhibitory cross-talk between car and HNF-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1α. J. Biol. Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Pascual G., Fong A.L., Ogawa S., Gamliel A., Li A.C., Perissi V., Rose D.W., Willson T.M., Rosenfeld M.G., Glass C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Ponugoti B., Fang S., Kemper J.K. Functional interaction of hepatic nuclear factor-4 and proliferator-activated receptor-γ coactivator 1α in CYP7A1 regulation is inhibited by a key lipogenic activator, sterol regulatory element-binding protein-1c. Mol. Endocrinol. 2007;21:2698–2712. doi: 10.1210/me.2007-0196. [DOI] [PubMed] [Google Scholar]
- Russell D.W. Nuclear orphan receptors control cholesterol catabolism. Cell. 1999;97:539–542. doi: 10.1016/s0092-8674(00)80763-1. [DOI] [PubMed] [Google Scholar]
- Russell D.W., Setchell K.D.R. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Bengoechea-Alonso M.T., Ye X., Lukiyanchuk V., Jin J., Harper J.W., Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Wang L., Lee Y., Bundman D., Han Y., Thevananther S., Kim C., Chua S., Wei P., Heyman R., Karin M., et al. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D.D. The orphan nuclear receptor SHP regulates PGC-1α expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Wang L., Huang J., Saha P., Kulkarni R.N., Hu M., Kim Y.D., Park K.G., Chan L., Rajan A.S., Lee I., et al. Orphan receptor small heterodimer partner is an important mediator of glucose homeostasis. Mol. Endocrinol. 2006;20:2671–2681. doi: 10.1210/me.2006-0224. [DOI] [PubMed] [Google Scholar]
- Xu Z., Tavares-Sanchez O.L., Li Q., Fernando J., Rodriguez C.M., Studer E.J., Pandak W.M., Hylemon P.B., Gil G. Activation of bile acid biosynthesis by the p38 mitogen-activated protein kinase (MAPK): Hepatocyte nuclear factor-4α phosphorylation by the p38 MAPK is required for cholesterol 7α-hydroxylase expression. J. Biol. Chem. 2007;282:24607–24614. doi: 10.1074/jbc.M611481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Wang F., Jin C., Huang X., McKeehan W.L. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J. Biol. Chem. 2005;280:17707–17714. doi: 10.1074/jbc.M411771200. [DOI] [PubMed] [Google Scholar]