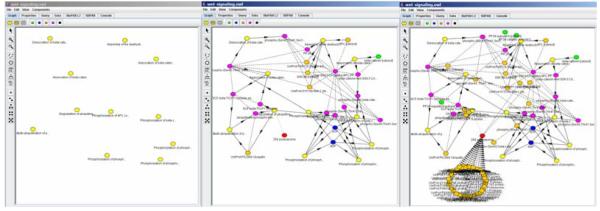
Figure 1.
The screenshot of BioPAX@VCell representation. The file with BioPAX file describing Signaling by Wnt [Homo Sapiens] was loaded from Reactome database. The pathway describes 11 interactions, but SBML file generated by Reactome includes about 100 species, to account for all proteins that constitute complexes in the reaction network. Visualization of this file in any simulation tool is unreadable. BioPAX@VCell provides a better visualization using two key features: (1) different coloring and symbols for different types of species and reactions; and (2) different level of granularity in representation, where the user can view, for example, reactions only (the first panel), reactions with reaction participants (second panel), or expansion of all complexes to view their components (the third panel). The user is able to collapse selected portions of the network into a single super-node, thus significantly improving network readability.