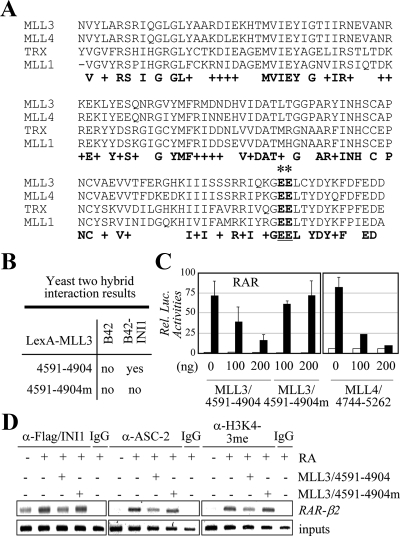
Figure 5.
Suppression of RAR transactivation by MLL3/4-SET involves their interactions with INI. A, The SET domains of MLL3, MLL4, TRX, and MLL1 are as shown. Asterisks indicate two residues originally described in TRX to be involved with recognizing INI1 (47). B, The yeast two-hybrid results for LexA-MLL3/4591-4904 and LexA-MLL3/4591-4904m and B42 alone and B42-INI1. C, The impact of MLL3/4591-4904, MLL3/4591-4904m, and MLL4/4744-5262 on RAR transactivation was measured using RARE-LUC reporter in HeLa cells. D, HEK293 cells transfected with Flag-INI1, either without or with MLL3/4591-4904 or MLL3/4591-4904m, were subjected to ChIP analyses for the recruitment of INI1 and ASC-2 to RAR-β2 as well as for H3K4 trimethylation of RAR-β2. Before ChIP, cells were treated with 0.1 μm RA for 15 min (for INI1 and ASC-2) or 1 h (for H3K4 trimethylation).