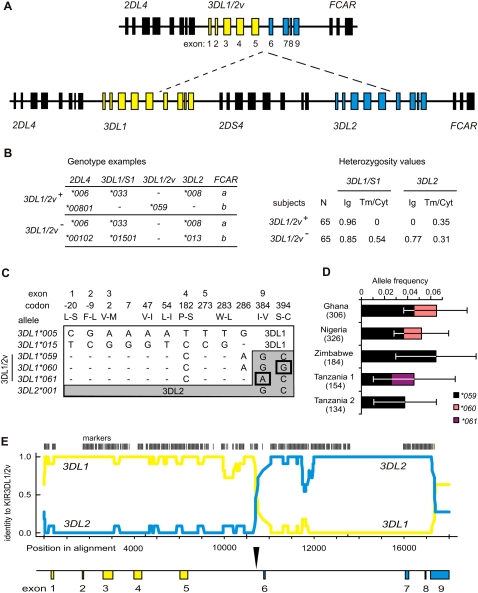
Figure 1.
The 3DL1/2v fusion gene is allelic to 3DL1/S1 and 3DL2. (A, top) Schematic of the four 3DL1/2v haplotypes that were sequenced here (Supplemental Fig. S2). (Bottom) A KIR A haplotype; (dashed lines) the genomic segment absent from 3DL1/2v haplotypes; (yellow) 3DL1; (blue) 3DL2. Exons 1–5 encode the leader peptide and Ig domains (Ig), exon 6 the stalk, and exons 7–9 the transmembrane and cytoplasmic domains (Tm/Cyt). (B, left) Representative genotypes from the group of 65 subjects who carry 3DL1/2v (3DL1/2v+) and the group of 65 who do not (3DL1/2v−). (Right) Comparisons of heterozygosity observed in the 3DL1/2v+ and 3DL1/2v− groups from segments of 3DL1 and 3DL2. All of the 3DL1/2v+ subjects are hemizygous for exons 6–9 of 3DL1/S1 and exons 1–5 of 3DL2, which corresponds to the portion absent from the KIR A haplotype. This shows the 3DL1/2v+ subjects have 3DL1 and 3DL2 on one haplotype and 3DL1/2v on the other. (C) Shown are the nucleotide differences in exons 1–5 that distinguish 3DL1/2v (3DL1*059, 3DL1*060, and 3DL1*061) from (white) 3DL1 and in exons 7–9 that distinguish 3DL1/2v from (gray) 3DL2. The many nucleotide differences (N = 102) that distinguish 3DL1 and 3DL2 are not shown. Shown are 3DL1*00501 and 3DL1*01502, which represent the 005 and 015 lineages of inhibitory receptors; exons 1–5 of 3DL1/2v are related to 3DL1*00501. In exons 7–9, 3DL1*059 is identical to 3DL2*001; 3DL1*060 and 3DL1*061 being distinguished by the SNPs boxed. Codons are numbered according to the mature protein, and amino acid changes are indicated by single letter code. (D) Shown are the frequencies of three 3DL1/2v alleles (3DL1*059–61) in the five sub-Saharan African populations where they were detected. The number of haplotypes examined from each population is shown in parentheses; error bars show the 95% confidence interval of the allele frequency measurements. (E) Shown is a pairwise identity plot from alignment of genomic sequences. (Yellow line) Signifies identity of 3DL1/2v with 3DL1; (blue line) the identity of 3DL1/2v with 3DL2; (vertical bars) the SNP markers used in this analysis. (Shown below by the vertical arrow) The crossover occurred in intron 5 during the interval from 386 to 356 bp upstream from exon 6. A continuous sequence trace that spans the crossover is shown in Supplemental Figure S2.