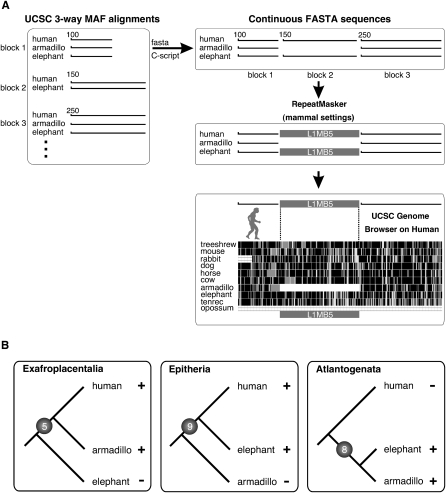
Figure 2.
Computational strategy to extract phylogenetically informative retroposon presence/absence loci. (A) UCSC three-way MAF data were organized in conserved alignment blocks with chromosomal coordinates. Three adjacent blocks including an internal gap region in block 2 were transformed in FASTA format. Sequences above gap regions were analyzed for mammalian specific repeats using the RepeatMasker. Loci including such repeats were inspected in the UCSC Genome Browser. All available sequences of phylogenetically informative loci were transformed to FASTA sequences and manually realigned. (B) Five, nine, and eight loci were identified that support the three competing hypotheses of the origin of placental mammals, Exafroplacentalia, Epitheria, and Atlantogenata, respectively. (+) Presence of a diagnostic retroposon; (−) absence of a diagnostic retroposon.