Table 2.
Chimpanzee-specific divergent sites seen in previous studies but not replicated in our analysis
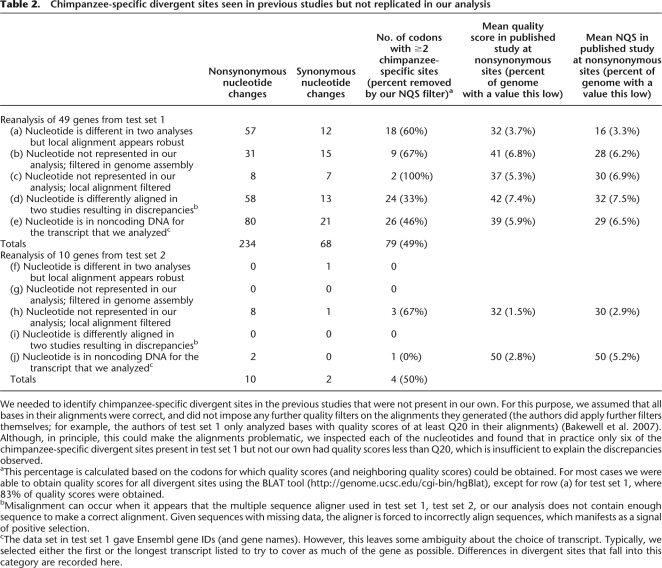
We needed to identify chimpanzee-specific divergent sites in the previous studies that were not present in our own. For this purpose, we assumed that all bases in their alignments were correct, and did not impose any further quality filters on the alignments they generated (the authors did apply further filters themselves; for example, the authors of test set 1 only analyzed bases with quality scores of at least Q20 in their alignments) (Bakewell et al. 2007). Although, in principle, this could make the alignments problematic, we inspected each of the nucleotides and found that in practice only six of the chimpanzee-specific divergent sites present in test set 1 but not our own had quality scores less than Q20, which is insufficient to explain the discrepancies observed.
aThis percentage is calculated based on the codons for which quality scores (and neighboring quality scores) could be obtained. For most cases we were able to obtain quality scores for all divergent sites using the BLAT tool (http://genome.ucsc.edu/cgi-bin/hgBlat), except for row (a) for test set 1, where 83% of quality scores were obtained.
bMisalignment can occur when it appears that the multiple sequence aligner used in test set 1, test set 2, or our analysis does not contain enough sequence to make a correct alignment. Given sequences with missing data, the aligner is forced to incorrectly align sequences, which manifests as a signal of positive selection.
cThe data set in test set 1 gave Ensembl gene IDs (and gene names). However, this leaves some ambiguity about the choice of transcript. Typically, we selected either the first or the longest transcript listed to try to cover as much of the gene as possible. Differences in divergent sites that fall into this category are recorded here.