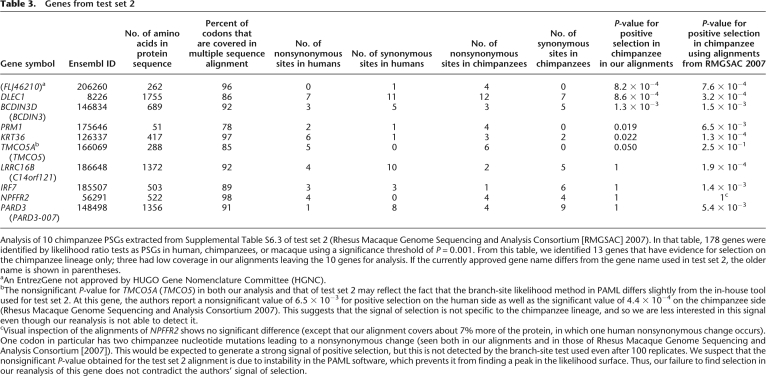
Table 3.
Genes from test set 2
Analysis of 10 chimpanzee PSGs extracted from Supplemental Table S6.3 of test set 2 (Rhesus Macaque Genome Sequencing and Analysis Consortium [RMGSAC] 2007). In that table, 178 genes were identified by likelihood ratio tests as PSGs in human, chimpanzees, or macaque using a significance threshold of P = 0.001. From this table, we identified 13 genes that have evidence for selection on the chimpanzee lineage only; three had low coverage in our alignments leaving the 10 genes for analysis. If the currently approved gene name differs from the gene name used in test set 2, the older name is shown in parentheses.
aAn EntrezGene not approved by HUGO Gene Nomenclature Committee (HGNC).
bThe nonsignificant P-value for TMCO5A (TMCO5) in both our analysis and that of test set 2 may reflect the fact that the branch-site likelihood method in PAML differs slightly from the in-house tool used for test set 2. At this gene, the authors report a nonsignificant value of 6.5 × 10−3 for positive selection on the human side as well as the significant value of 4.4 × 10−4 on the chimpanzee side (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). This suggests that the signal of selection is not specific to the chimpanzee lineage, and so we are less interested in this signal even though our reanalysis is not able to detect it.
cVisual inspection of the alignments of NPFFR2 shows no significant difference (except that our alignment covers about 7% more of the protein, in which one human nonsynonymous change occurs). One codon in particular has two chimpanzee nucleotide mutations leading to a nonsynonymous change (seen both in our alignments and in those of Rhesus Macaque Genome Sequencing and Analysis Consortium [2007]). This would be expected to generate a strong signal of positive selection, but this is not detected by the branch-site test used even after 100 replicates. We suspect that the nonsignificant P-value obtained for the test set 2 alignment is due to instability in the PAML software, which prevents it from finding a peak in the likelihood surface. Thus, our failure to find selection in our reanalysis of this gene does not contradict the authors' signal of selection.