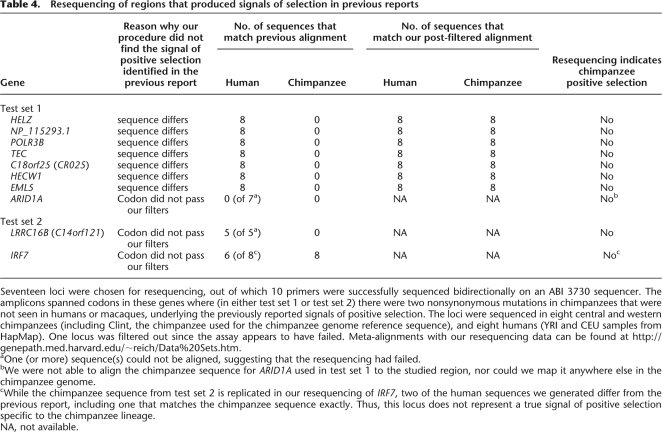
Table 4.
Resequencing of regions that produced signals of selection in previous reports
Seventeen loci were chosen for resequencing, out of which 10 primers were successfully sequenced bidirectionally on an ABI 3730 sequencer. The amplicons spanned codons in these genes where (in either test set 1 or test set 2) there were two nonsynonymous mutations in chimpanzees that were not seen in humans or macaques, underlying the previously reported signals of positive selection. The loci were sequenced in eight central and western chimpanzees (including Clint, the chimpanzee used for the chimpanzee genome reference sequence), and eight humans (YRI and CEU samples from HapMap). One locus was filtered out since the assay appears to have failed. Meta-alignments with our resequencing data can be found at http://genepath.med.harvard.edu/~reich/Data%20Sets.htm.
aOne (or more) sequence(s) could not be aligned, suggesting that the resequencing had failed.
bWe were not able to align the chimpanzee sequence for ARID1A used in test set 1 to the studied region, nor could we map it anywhere else in the chimpanzee genome.
cWhile the chimpanzee sequence from test set 2 is replicated in our resequencing of IRF7, two of the human sequences we generated differ from the previous report, including one that matches the chimpanzee sequence exactly. Thus, this locus does not represent a true signal of positive selection specific to the chimpanzee lineage.
NA, not available.