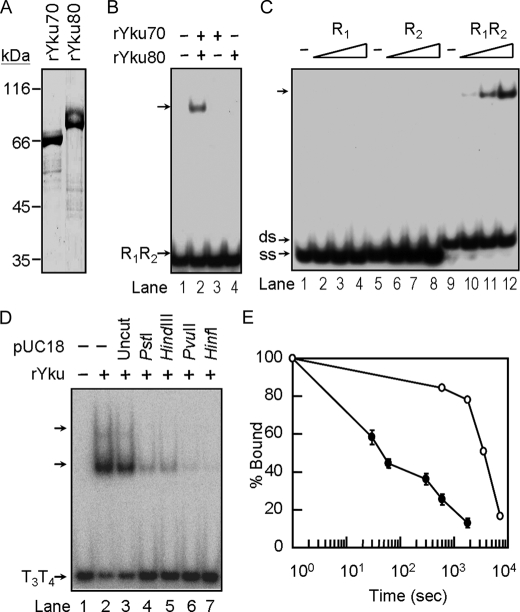
FIGURE 5.
Stably formed Yku is required for ternary complex formation. A, purification of Yku70 and Yku80 from insect cells. Yku70 and Yku80 with His6 tag were purified from Sf21 using a Ni-NTA-agarose column, respectively (see “Experimental Procedures”). Five μg each of purified Yku70 and Yku80 were analyzed on a 10% SDS-polyacrylamide gel and stained with Coomassie Blue, respectively. B, Yku heterodimer is essential for DNA binding activity. Yku70 and/or Yku80 of 100 nm was incubated with 2 ng of radiolabeled R1R2 DNA duplex and analyzed by EMSA. An autoradiogram is presented. C, reconstituted Yku binds to duplex DNA. 10, 33, or 100 nm of reconstituted Yku was incubated with 2 ng of radiolabeled oligonucleotide R1, R2, or R1R2 duplex DNA and then analyzed by EMSA. An autoradiogram is presented. D, Yu binds to the ends of the tailed-duplex DNA. Around 2 ng each of the tailed-duplex DNA formed by T3 and T4 were first mixed without (lanes 1 and 2) or with 200 ng of pUC18 plasmid DNA that was undigested (lanes 3) or digested by PstI (1 cut, lanes 4), HindIII (1 cut, lane 5), PvuII (2 cuts, lane 6), or HinfI (6 cuts, lanes 7). The DNA mixtures were then incubated with 150 nm of Yku and then analyzed by electrophoresis using a 6% polyacrylamide gel. An autoradiogram is shown here. E, DNA binding stability of Yku. Yku of 100 nm isolated from yeast (open circle) or reconstituted Yku (close circle) were incubated with 0.2 ng of radiolabeled R1R2 at room temperature for 10 min. Twenty ng of unlabeled R1R2 were added to the reaction mixtures. At the time indicated, an aliquot of the reaction mixtures was withdrawn and analyzed by EMSA. The amounts of DNA remained bound by Yku were quantified by a PhosphorImager. The value obtained at time = 0 was taken as 100%. The data show the average of three experiments.