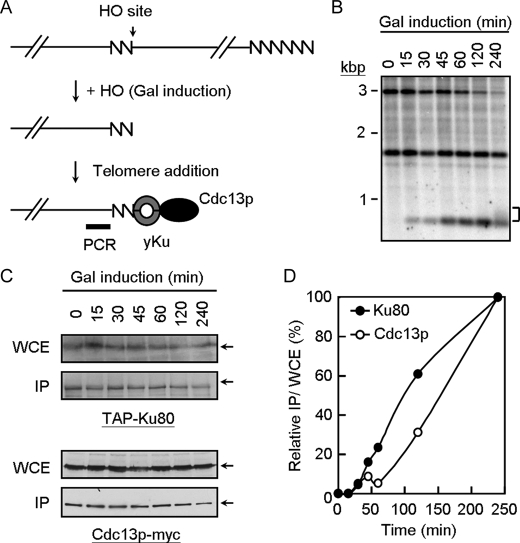
FIGURE 7.
Loading of Yku to telomeres is earlier than that of Cdc13p. A, schematic representation of the de novo telomere addition assay (32). The 81 bp of TG1–3 sequence (zigzag line) and the recognition site for the HO endonuclease was placed near the telomere of chromosome VII-L. HO is induced by the addition of galactose to the media. The cells then add TG1–3 to the TG1–3/HO end to repair the end. The probe used to monitor the TG1–3/HO end is shown as a thick bar. Yeast cells (YJL0801) were treated with nocodazole, transferred into medium containing galactose to induce HO expression, and then samples were taken at the time indicated. B, Southern blot analysis of SpeI cut genomic DNA is shown. The band labeled at ∼3 kbp is the SpeI fragment from the construct on chromosome VII-L. After cleavage with HO, this fragment is converted into a new band with the size of ∼0.7 kbp. The 1.6-kbp SpeI fragment represents the endogenous ade2-101 locus is also indicated. This band serves as a DNA loading control. Bracket indicates the newly synthesized telomeres from the 0.7-kbp fragment. C, immunoblotting analysis of the total yeast extracts and immunoprecipitates (IP) using antibodies against TAP or Cdc13p. WCE, whole cell extract. D, real time PCR analysis of the immunoprecipitated DNA. The results from real time PCR experiments are expressed as the relative value of immunoprecipitates/input using time = 0 as 0% and time = 240 min as 100%. The values presented are the average of 3–4 independent experiments.