Abstract
γ-Secretase is a multisubunit membrane protein complex consisting of presenilin (PS1), nicastrin (NCT), anterior pharynx-1, and presenilin enhancer 2. To analyze the activity of familial Alzheimer disease mutants and to understand the roles of the subunits, we established a yeast transcriptional activator Gal4p system with artificial γ-secretase substrates containing amyloid precursor protein or Notch fragments. The γ-secretase activities were evaluated by transcriptional activation of reporter genes upon Gal4p release from the membrane-bound substrates, i.e. growth of yeast on histidine and adenine, or β-galactosidase assay. We screened and evaluated γ-secretase mutants using this reconstitution system in yeast, which does not possess endogenous γ-secretase activity. When we introduced familial Alzheimer mutants of PS1 in this system, their activities were shown to be loss of function. Although the protease activity of wild type PS1 depends on the other three subunits introduced, we obtained 15 new PS1 mutants, which are active in the absence of NCT. They possessed a S438P mutation at the ninth transmembrane domain (TM9) together with one missense mutation distributed through transmembrane and loop regions. These mutations were not related to familial Alzheimer mutations of PS1 as identified so far. The S438P mutant was partially active but required other mutations for full activation. Results of the β-galactosidase assay suggested that they have wild type protease activities, which were further confirmed by the endoproteolysis of PS1, amyloid β peptides, and Notch intracellular domain production in mammalian cells. These results suggest that NCT is dispensable for the protease activity of γ-secretase.
γ-Secretase mediates an intramembrane cleavage of type I integral membrane proteins, including amyloid precursor protein (APP)2 and Notch. Abnormal processing of APP produces a small amyloid β fragment (Aβ42), possibly responsible for Alzheimer disease (1). γ-Secretase is composed of four membrane proteins as follows: presenilin (PS; PS1 or PS2), nicastrin (NCT), anterior pharynx-1 (Aph-1), and presenilin enhancer-2 (Pen2), which are necessary for the protease activity (2). PS contains nine transmembrane domains (TM1 through TM9) (3–5), whereas Aph-1, Pen2, and NCT contain seven, two, and one, respectively. PS is believed to be the subunit with aspartyl protease activity (6). More than 100 human missense mutations in PS (1, 7) increased the production of Aβ42 peptides, and they are associated with early onset familial Alzheimer disease (FAD). NCT interacts with the luminal region of the APP fragments, leading to the hypothesis that NCT functions as a substrate acceptor (8). On the other hand, Pen2 triggers the endoproteolysis of PS into amino- and carboxyl-terminal fragments (called NTF and CTF, respectively) as a part of the maturation of the protease complex (9). Aph-1 is thought to be a scaffold for the assembly and contributes to the stability of the entire complex and its trafficking to the Golgi apparatus (10).
The interactions between the subunits and the arrangement of PS1 transmembrane domains were partly understood from biochemical analyses. The carboxyl-terminal region (including TM8 and TM9) and TM4 of PS1 interact with NCT (or Aph-1) (11–13) and Pen2 (14, 15), respectively. In addition to the substrate-binding site in NCT, Kornilova et al. (16) proposed that PS1 contains a substrate recognition and a catalytic site. Prior to the entry into the catalytic site, the substrate interacts with the recognition site (also called docking site), which may be identical to the binding sites of APP and telencephalin (TM1 and the carboxyl-terminal region containing TM9) (17). The catalytic site of PS1 was further analyzed by cysteine scanning mutagenesis; TM6 and TM7 form water-containing cavities inside the membrane with two functional Asp residues (Asp257 and Asp385, respectively (6)) located very closely (18, 19). The TM9 and a hydrophobic domain between TM6 and TM7 were also shown to be nearby (20, 21).
The structural arrangements of γ-secretase subunits were known at least partially (22). However, the catalytic mechanism has yet to be understood, including the roles of NCT, Aph-1, and Pen2 to trigger protease activity. Signal peptide peptidases (SPP and SPP-like proteases) were found to be structural homologues of PS (23, 24). They have two conserved Asp residues for the aspartyl protease activity (23), and cleave membrane-embedded signal peptides (25) or type II membrane proteins (24). SPP shares a series of inhibitors with γ-secretase (25, 26), suggesting that SPP and PS have similar catalytic sites. Although they are similar in protease activity, SPP does not require additional subunit(s) for activity (27). Thus, γ-secretase subunits, NCT, Aph-1, and Pen2 may not be required for proteolysis itself. To address this question, it is of interest to test whether mutations in PS1 can suppress the lack of other subunits.
Edbauer et al. (2) indicated that the four subunits are essential for protease activity using the yeast transcriptional activator Gal4 system with artificial γ-secretase substrate (C1–55-Gal4p), which contains APP fragment. We extended this system for analyzing the FAD mutants of PS1 and the roles of γ-secretase subunits. We screened and evaluated γ-secretase mutants in yeast, which does not possess functional homologues of the protease complex. PS1 FAD mutants were shown to be loss of function in the yeast system similar to mammalian cells. Furthermore, we isolated 15 new PS1 mutants, which do not require NCT to proteolyze the Gal4-fused substrates and endoproteolyze PS1 itself in the yeast system. They contain a common S438P mutation in TM9, together with one missense mutation found in TM1, TM3, TM5, TM6, TM8, TM9, and loop regions. These mutations were not related to the FAD mutations of PS1. Using mouse embryonic fibroblasts with PS knock-out (28, 29) or NCT knock-out (30, 31), we could show that these mutant PS1s becomes mature form by endoproteolysis, producing Aβ and Notch intracellular domain (NICD) in the absence of NCT. These observations suggest that PS1 with the critical S438P mutation in TM9 does not require the substrate receptor NCT for the protease activity.
EXPERIMENTAL PROCEDURES
γ-Secretase and APP Constructs—The expression constructs for γ-secretase were prepared as described (2) with minor modifications. PS1 and NCT were cloned into KpnI and XbaI sites of pBEVY-T vector (32), respectively. FLAG-Pen2 and Aph-1-HA (Aph-1aL splice variant) were cloned into KpnI and XbaI sites of pBEVY-L vector (32), respectively. PS1 and FLAG-Pen2 were expressed by the ADH1 promoter, and NCT and Aph-1 were expressed by the GPD promoter. APP-based substrate, C1–55-Gal4p, and Notch-based substrate, NotchTM-Gal4p, were prepared; DNA fragment for C1–55 (amino acids 672–726 of the human APP770 isoform) or NotchTM (amino acids 1703–1754 of the mouse Notch-1) was amplified by PCR using primers encoding a 19-amino acid signal peptide sequence from yeast invertase (SUC2) to make SUC2-C1–55 or SUC2-NotchTM. They were ligated with the GAL4 gene and cloned into BamHI and EcoRI sites of p426ADH (33). DNA for C1–99 (amino acids 672–770 of the human APP770) was also amplified by PCR and cloned into BamHI site of p426ADH (33) to express C1–99 fragment in yeast. For expression in mammalian cells, wild type or mutant PS1 was inserted at the KpnI site of pcDNA3.1/Zeo to make PS1pcDNA3.1/Zeo.
Yeast Transformation and Assay for Reporter Gene Expression—Recombinant plasmids were transformed into Saccharomyces cerevisiae strain PJ69-4A (MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) (34). Transformants were grown on SD agar medium lacking Leu, Trp, and Ura (SD-LWU). The expressions of HIS3 (His) and ADE2 (Ade) were assessed by colony growth on selection medium plate, SD-LWHUAde. β-Galactosidase was assayed at 30 °C using o-nitrophenyl β-d-galactopyranoside, as described previously (35). Exponentially grown cells (1 × 107 cells) were lysed by glass beads in 30 μl of lysis buffer (20 mm Tris/Cl (pH 8.0), 10 mm MgCl2, 50 mm KCl, 1 mm EDTA, 5% glycerol, 1 mm dithiothreitol) including protease inhibitor mixture (Sigma). Cell lysate was centrifuged for 10 min at 15,000 × g, and the supernatant was used to determine β-galactosidase activity and protein concentration (the Bradford protein assay, Bio-Rad).
Random Mutagenesis by PCR—Random mutations were introduced using human PS1 cDNA and following two primers: PS1S, 5′-TTCAAGCTATACCAAGCATACAATCAACTCCCCGGGTACCAAAAATGACAGAGTTACCTGCACCGTTG-3′, and PS1AS, 5′-GATCCGCTTATTTAGAAGTGTCGAATTCGACCTCGGTACCATGCTAGATATAAAATTGATGGAATGC-3′. PCR was performed in 50 μl of the solution (50 ng of template DNA, 0.2 mm dGTP, 1 mm dATP/dTTP/dCTP, 400 nm each primer, 3 mm MgCl2, 0.5 mm MnCl2 in 1× rTaq buffer) with 1.25 units of rTaq using the following cycles: 94 °C for 5 min; 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 120 s for 30 cycles; 72 °C for 10 min. The error ratio of this condition was about 2.6 mutations per PS1 gene. The mutagenized PS1 cDNA fragments (∼4 μg) were cotransformed with 4 μg of the KpnI fragment of pBEVY-T into PJ69-4A. The PS1 primers, PS1S and PS1AS, contain 40-bp regions from pBEVY-T (32) at the 5′ termini, which enable ligation in vivo by homologous recombination (36). About 2 × 105 transformants were screened on selection medium plates, SD-LWHUAde (Table 1). Plasmid DNAs were isolated from yeast colonies, and mutations were identified by DNA sequencing. We also introduced site-directed mutations by Quick-Change mutagenesis kit (Stratagene).
TABLE 1.
Screening for PS1 mutants active without other subunits
| Other subunit | No. of cells tested | Positive clones | False positives |
|---|---|---|---|
| Pen2 | 3.3 × 105 | 0 | 18 |
| Aph-1 | 2.4 × 105 | 0 | 10 |
| NCT | 1.9 × 105 | 1 | 11 |
Yeast Microsome Preparations for Immunoblotting or γ-Secretase Assay—Yeast microsomes were prepared as described previously (37). Microsomal membranes were suspended in γ-buffer (50 mm PIPES (pH 7.0), 250 mm sucrose, 1 mm EGTA) and subjected to immunoblotting or γ-secretase assay (38). The γ-secretase activity was analyzed after solubilization as follows: microsomes (40 μg) were solubilized in the presence of 1% CHAPSO on ice for 1 h, and diluted 4-fold with γ-buffer containing protease inhibitors (PI mix) to lower the CHAPSO concentration. PI mix contained the following: 50 μm diisopropyl fluorophosphate, 50μm phenylmethylsulfonyl fluoride, 100 ng/ml Nα-p-tosyl-l-lysine chloromethyl ketone, 100 ng/ml antipain, 100 ng/ml leupeptin, 1 mm thiorphan, 5 mm phenanthroline monohydrate, and 0.1 mm EGTA. The mixture was incubated at 37 °C, and the protein fraction was recovered after chloroform/methanol (2:1) extraction (39). The Aβ production was analyzed by immunoblotting after gel electrophoresis (39). Intensities of the signals were quantified using an LAS-3000 luminescent image analyzer (Fuji Film, Tokyo, Japan).
Immunoprecipitation of γ-Secretase—Yeast microsomes (80 μg) were solubilized with the IP buffer (1% CHAPSO, 50 mm Hepes (pH 7.4), 150 mm NaCl, 2 mm EDTA, with the protease inhibitor mixture (Sigma)) on ice for 1 h and centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant was incubated with primary antibody (dilution 1:300) overnight at 4 °C and incubated with protein A-Sepharose beads for 1 h at room temperature (GE Healthcare). Then the immunoprecipitates on beads were washed with the IP buffer and subjected to immunoblotting.
Protein Expression and siRNA Transformation in Mouse Embryonic Fibroblasts—Mouse embryonic fibroblast cell with PS1/PS2 double knock-out (28, 29) was transfected with PS1pcDNA3.1/Zeo constructs, C1–99pcDNA (40), and Stealth RNA interference for mouse nicastrin (MSS226913, Invitrogen) or Stealth RNA interference negative control (Medium GC, Invitrogen) using Lipofectamine 2000 (Invitrogen). Mouse embryonic fibroblast cell with NCT knock-out (30, 31) was transfected with PS1pcDNA3.1/Zeo constructs, and C1–99pcDNA (40) or pCS2+mNΔE myc (41), using Lipofectamine 2000. After 48 h of incubation, cells and the media were recovered and subjected to immunoblotting. The knock-out cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Antibodies—The antibodies used include the following: monoclonal antibodies against Aβ (82E1) (IBL, Fujioka, Japan); HA (12CA5) (Sigma); and FLAG (M2) (Sigma) and the polyclonal antibodies against APP carboxyl terminus (A8717) (Sigma); NCT (AB5890) (Chemicon); cleaved Notch intracellular domain (Val1744) (Cell Signaling); PS1 amino terminus (G1Nr3); PS1 loop region (G1L3) (the gift from Dr. T. Iwatsubo and Dr. T. Tomita, University of Tokyo) (42); and PS1 loop region (PS1L) (43) (the gift from Dr. T. Saido).
RESULTS
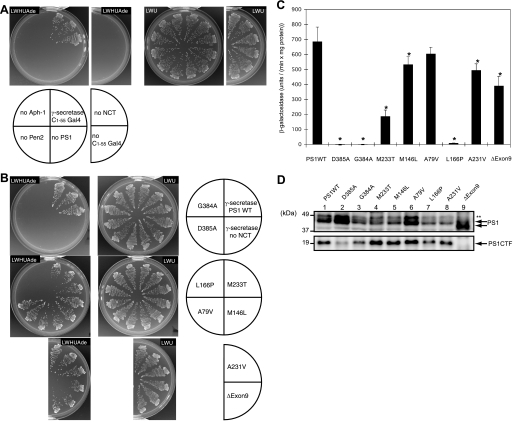
Effects of Familial Alzheimer Disease Mutations on γ-Secretase Activity—We have constructed recombinant plasmids for γ-secretase and APP-based substrate (C1–55-Gal4p) and introduced them into yeast strain PJ69-4A, which possesses HIS3, ADE2, and lacZ marker under Gal4p control. Gal4p cleaved from the membrane-bound C1–55-Gal4p by γ-secretase activates the transcription of HIS3 and ADE2 genes. Thus, the protease activity could be monitored by the positive cell growth in the media lacking histidine and adenine. Cells could grow with four subunit genes introduced but could not when lacking any one of them (Fig. 1A), indicating that they are essential for the γ-secretase activity.
FIGURE 1.
Reconstitution of PS1 FAD mutants in yeast. A and B, γ-secretase subunits were introduced into yeast with C1–55-Gal4p. Gal4p cleaved from C1–55-Gal4p activates the reporter genes, HIS3 and ADE2, and the release was assessed by the growth on the selection medium, lacking histidine and adenine. Cells expressing PS1 wild type (WT)(A) or FAD mutants (B), other γ-secretase subunits, and C1–55-Gal4p were examined for growth after 3 days at 30 °C on selection (SD-LWHUAde) or nonselection (SD-LWU) media, as indicated. Three independent clones were tested for each strain. C, β-galactosidase activity was measured for each yeast strain with PS1 wild type or FAD mutants, as indicated. One unit of β-galactosidase activity corresponds to 1 nmol of o-nitrophenyl β-d-galactopyranoside hydrolyzed per min, and activity was calculated as unit/(min × mg of protein in lysate). The activity was normalized by the subtraction with the activity in the absence of NCT, 38.7 unit/(min × mg of protein). Representative results from three independent assays are shown with the standard deviations. Statistical analyses were performed by one-way analysis of variance followed by Dunnett's multiple comparison test. Asterisks indicate p < 0.01 with respect to PS1 wild type. D, expression of wild type and PS1 mutants in lysates were analyzed by immunoblotting using antibody against PS1 (G1L3). Arrows indicate full-length PS1 and CTF (carboxyl-terminal fragment). ΔExon9 mutant migrates faster than the wild type. ** indicates nonspecific bands.
We introduced familial Alzheimer disease (FAD) mutations in the PS1 gene and transformed them into yeast together with three other subunits. Cells with G384A or L166P mutants could not grow on media lacking histidine and adenine similar to an active site mutant D385A (Fig. 1B). On the other hand, exon 9 deletion (ΔExon9) or M233T mutant could grow, although their rates were apparently slower than wild type (Fig. 1B). β-Galactosidase activity could semi-quantitatively determine the amount of Gal4p released by γ-secretase; the β-galactosidase activities could be roughly correlated with the growth phenotype (Fig. 1C), suggesting that the mutants possess lower protease activities than wild type. Immunoblotting confirmed that the expression of mutant PS1s was at a similar level (Fig. 1D). The mutants, M233T, M146L, A79V, A231V, and ΔExon9, required subunits (NCT, Aph-1, and Pen2) for growth (data not shown).
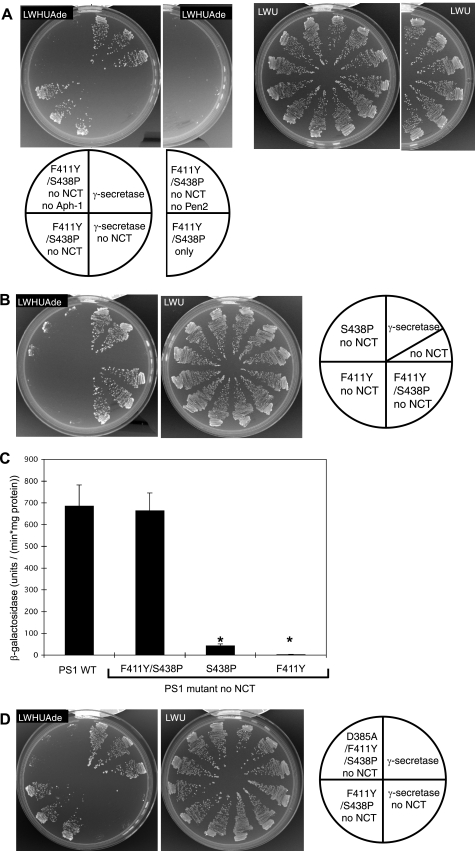
PS1 Mutant S438P/F411Y Is Active in the Absence of NCT—It became of interest to isolate PS1 mutants able to grow in the absence of NCT, Aph-1, or Pen2 (Table 1). After screening a number of cells having PS1 mutations, one mutant was found to grow in the absence of NCT (Fig. 2A). The mutant still required Aph-1 and Pen2 for growth (Fig. 2A) and possesses two missense mutations, F411Y and S438P, mapped at TM8 and TM9 transmembrane segments, respectively. When we prepared cells with each replacement, F411Y did not grow and S438P grew partially (Fig. 2B), indicating that the two mutations are necessary for full activation of growth. β-Galactosidase activity showed that the F411Y/S438P mutant possesses wild type activities in the absence of NCT (Fig. 2C). The S438P single mutant had slight β-galactosidase activity (Fig. 2C), suggesting the partial activation. These results suggest that γ-secretase with PS1 F411Y/S438P mutation can be active in the absence of NCT.
FIGURE 2.
PS1 mutant, F411Y/S438P, active in the absence of NCT. A and B, cells expressing PS1 wild type, F411Y/S438P (A), F411Y, or S438 (B) mutants, otherγ-secretase subunits, and C1–55-Gal4p were examined for growth after 3 days at 30 °C on selection (SD-LWHUAde) or nonselection (SD-LWU) media, as indicated. C, β-galactosidase activity was measured for each yeast strain containing wild type or mutant PS1, as indicated. The experimental conditions are as in Fig. 1C. Data represent means ± S.D.;n=3. Asterisks indicate p < 0.01 with respect to PS1 wild type. D, cells expressing D385A/F411Y/S438P, the catalytic site mutant, were examined for growth as indicated.
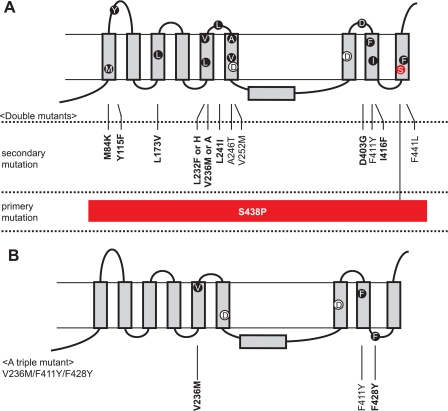
S438P Mutation Is Important for the PS1 Activation—To further assess the PS1 mutation, we have replaced Phe411 and Ser438 residues randomly. None of the possible combinations gave the same growth as the original F411Y/S438P (supplemental Table 1). However, single S438P replacement or those with F411G, F411P, F411A, F411I, F411M, and F411V could grow slightly but significantly on SD-LWHUAde media, suggesting that they are partially active without NCT (supplemental Table 1). To obtain more mutants similar to F411Y/S438P, we further screened mutants by introducing random mutations into PS1 having F411Y or S438P replacement (Table 2). We obtained 41 and 47 positive clones from F411Y and S438P, respectively. 40 of those from F411Y were the original F411Y/S438P mutants (Table 2 and Fig. 3A). One exception had three replacements V236M/F428Y/F411Y (Fig. 3B). These results indicate that the S438P is pertinent for the activity of the PS1 mutants. Starting from S438P, we obtained various replacements localized in TM1, TM3, TM5, TM6, TM8, and TM9, and loop regions (Table 2 and Fig. 3A). These mutations did not have consistent change in amino acid residues such as from acidic to neutral or from hydrophobic to hydrophilic, suggesting that the secondary mutations are less critical than S438P.
TABLE 2.
Screening for PS1 mutants active without NCT
| Mutants | No. of positive clones |
|---|---|
| Screening (3.7 × 105 cells tested) from PS1 F411Y mutant | |
| F411Y/S438P (the original mutant) | 40 |
| V236M/F428Y/F411Y | 1 |
| Screening (3.7 × 105 cells tested) from PS1 S438P mutant | |
| M84K/S438P | 1 |
| Y115F/S438P | 2 |
| L173V/S438P | 1 |
| L232H/S438P | 2 |
| L232F/S438P | 1 |
| V236A/S438P | 4 |
| V236M/S438P | 13 |
| L241I/S438P | 1 |
| A246T/S438P | 8 |
| V252M/S438P | 1 |
| D403G/S438P | 2 |
| F411Y/S438P (the original mutant) | 3 |
| I416F/S438P | 1 |
| F441L/S438P | 7 |
FIGURE 3.
PS1 mutants active in the absence of NCT. A and B, mutations in double mutants (A) or a triple mutant (B) are indicated in the model with nine transmembrane domains of PS1. All double mutants contained S438P as a primary mutation with a variable secondary mutation. Conserved residues are in boldface. Catalytic residues in TM6 and TM7 (Asp257 and Asp385) are also indicated.
We also replaced catalytic Asp385 in combination with F411Y/S438P. The mutant, D385A/F411Y/S438P, could not grow in the media lacking histidine and adenine (Fig. 2D), confirming that the growth phenotype of F411Y/S438P is because of the liberation of Gal4p by the mutant γ-secretase.
S438P/F411Y Mutant Could Process NotchTM-Gal4p in the Absence of NCT—To further test the specificity of mutant PS1, we tested another artificial γ-secretase substrate, NotchTM-Gal4p, which contains Notch-1 fragment. Without histidine or adenine, cells could grow only when four subunit genes were introduced but could not when lacking any one of them (Table 3 and supplemental Fig. 1), indicating that Gal4p was released only with four subunits of γ-secretase. However, cells with the F411Y/S438P mutant grew in the absence of NCT (Table 3 and supplemental Fig. 1). Their growth profiles were similar to cells with C1–55-Gal4p, suggesting that the F411Y/S438P mutant proteolyzes APP and Notch in the similar manner.
TABLE 3.
Processing of NotchTM-Gal4p by the F411Y/S438P mutant
| PS1 (wild type or mutant) | NCT | Aph-1 | Pen2 | Growth of cellsa |
|---|---|---|---|---|
| Wild type | + | + | + | +++ |
| Wild type | – | + | + | – |
| Wild type | + | – | + | – |
| Wild type | + | + | – | – |
| F411Y/S438P | + | + | + | +++ |
| F411Y/S438P | – | + | + | +++ |
| F411Y/S438P | + | – | + | – |
| F411Y/S438P | + | + | – | – |
| F411Y/S438P | – | – | + | – |
| F411Y/S438P | – | + | – | – |
| + | + | + | – |
The growth of cells with NotchTM-Gal4p and γ-secretase subunits was analyzed on SD-LWHUAde medium after 3 days at 30 °C. All cells contains NotchTM-Gal4p. +++ represents the full growth, cells formed colonies (>1 mm). – represents no growth
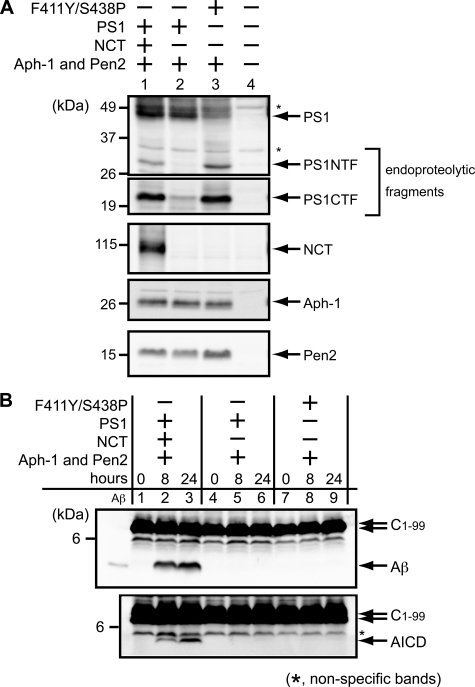
Endoproteolysis of Mutant PS1 in Yeast Cells—We analyzed γ-secretase endoproteolytic activity of PS1 mutant using the microsomal fraction. The endoproteolysis can be detected by the presence of NTF and CTF. The wild type PS1 with three other subunits gave two mature fragments, whereas it gave no fragments in the absence of NCT (Fig. 4A, lanes 1 and 2). On the other hand, the F411Y/S438P mutant produced the two endoproteolytic fragments in the absence of NCT (Fig. 4A, lane 3). The amounts of mutant fragments were similar to those of wild type with NCT (Fig. 4A, lanes 1 and 3). Those of other subunits, Aph-1 and Pen2, did not change with the PS1 mutation. Ratio of endoproteolytic fragments to uncleaved PS1 increased with the mutant (Fig. 4A, lanes 1 and 3), suggesting that the mutant had higher endoproteolytic activity than wild type with NCT.
FIGURE 4.
Endoproteolysis of F411Y/S438P mutant in yeast cells. A, microsomes were prepared from yeast transformants expressing PS1 wild type or PS1 F411Y/S438P mutant, NCT, Aph-1-HA, and FLAG-Pen2, as indicated. These subunits were analyzed by immunoblotting using specific antibodies. The endoproteolysis of PS1 was detected by the production of fragments, PS1 NTF and PS1 CTF. B, microsomes expressing γ-secretase and C1–99 fragment of APP were subjected to the γ-secretase assay. CHAPSO-solubilized microsomes were incubated at 37 °C for 0, 8, or 24 h. Aβ and AICD production was analyzed by immunoblotting using specific antibodies. Synthetic Aβ40 (30 pg) was loaded as a marker in the leftmost lanes. The asterisks indicate nonspecific bands.
Cleavage of APP C1–99 was tested in vitro by incubating the solubilized microsomes (38) (Fig. 4B). The two fragments, Aβ and the APP intracellular domain (AICD), were observed in the presence of γ-secretase with wild type PS1 (Fig. 4B, lanes 1–3). However, these fragments were not observed with wild type in the absence of NCT (Fig. 4B, lanes 4–6) or the mutants PS1 (Fig. 4B, lanes 7–9). We also tested the Aβ production using APP C1–55 fragment, but Aβ was not found in vitro with the F411Y/S438P mutant (data not shown).
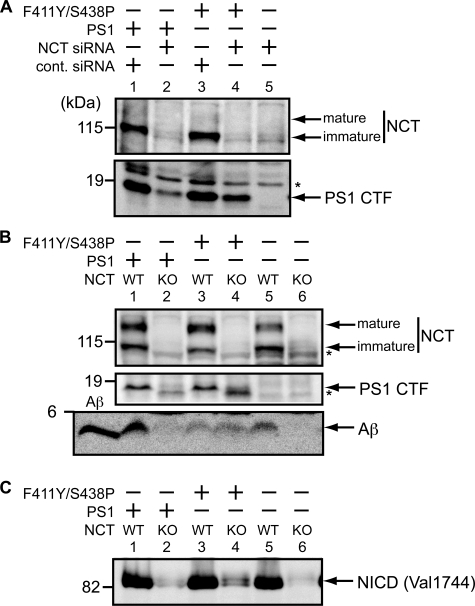
Endoproteolysis, Aβ, and NICD Production by Mutant PS1 in Mouse Fibroblasts—We have tested endoproteolytic activity of the PS1 mutant F411Y/S438P using a mouse embryonic fibroblast with PS1/PS2 double knock-out (Fig. 5A). NCT gene expression was blocked by the addition of NCT siRNA, and wild type or the mutant PS1 cDNA were transfected into the fibroblasts. The NCT knockdown was confirmed by immunoblotting (Fig. 5A, lanes 2 and 4). The mature fragments (CTF) were found in the presence of NCT with wild type or the mutant PS1 (Fig. 5A, lanes 1 and 3). In the knockdown cells, CTFs from wild type PS1 were barely detectable (Fig. 5A, lane 2), but significant amounts were observed from the mutant (Fig. 5A, lane 4), indicating that the F411Y/S438P mutant possesses endoproteolytic activity in mammalian cells. The full-length PS1s were not detected because they were under the detection level (data not shown).
FIGURE 5.
Endoproteolysis, Aβ, and NICD production by F411Y/S438P mutant in mouse embryonic fibroblasts. A, PS double knock-out cells were transiently transfected with C1–99pcDNA, PS1pcDNA3.1/Zeo, or F411Y/S438PpcDNA3. 1/Zeo, and siRNA for NCT or control siRNA (cont. siRNA), as indicated. Cells were recovered and analyzed by immunoblotting with specific antibodies. The endoproteolysis of PS1 was detected by the production of PS1 CTF. B, NCT knock-out (KO) cells or wild type (WT) fibroblast cells were transiently transfected with C1–99pcDNA, PS1pcDNA3.1/Zeo, or F411Y/S438PpcDNA3.1/Zeo as indicated. Cells and media were recovered and analyzed by immunoblotting with specific antibodies (human PS1-specific antibody (PS1L) was used for PS1). Aβ was detected in media. Synthetic Aβ40 (30 pg) was loaded as indicated. C, NCT knock-out cells (KO) or wild type (WT) fibroblast cells were transiently transfected with pCS2+mNΔEmyc, PS1pcDNA3.1/Zeo, or F411Y/S438PpcDNA3.1/Zeo as indicated. Membrane-bound mNotch-1 (NΔE) was expressed from the pCS2+mNΔEmyc vector. 48 h after transfection, cells were incubated with 10 μm lactacystin for 4 h, recovered, and analyzed by immunoblotting with specific antibodies. The asterisks indicate nonspecific bands.
Cleavage of C1–99 was tested in mouse embryonic fibroblasts with NCT knock-out (Fig. 5B). Aβ was found in the medium when all four subunits of γ-secretase were present (Fig. 5B, lanes 1 and 5), but it was not detected in the absence of NCT (Fig. 5B, lanes 2 and 6), confirming that NCT is required for the Aβ production. With the PS1 mutant, significant amounts of Aβ were found in the presence or in the absence of NCT (Fig. 5B, lanes 3 and 4), indicating that the F411Y/S438P mutant does not require NCT for Aβ production from C1–99. In the NCT knock-out cells, the maturation of the mutant PS1 was confirmed by the production of CTF (Fig. 5B, lane 4). The full-length PS1s were not detected (data not shown).
To further test the γ-secretase activity using another substrate, membrane-bound Notch-1 (NΔE (41)) was introduced in fibroblasts with NCT knock-out. NICD fragments were released from NΔE after the cleavage. NICD was found when all four subunits are present (Fig. 5C, lanes 1, 3, and 5). With the PS1 mutant, a significant amount of NICD was found even in the absence of NCT (Fig. 5C, lane 4), indicating that the F411Y/S438P mutant does not require NCT for Notch cleavage.
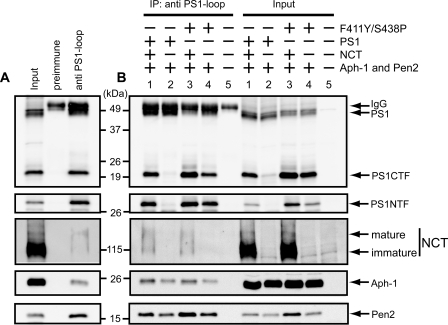
F411Y/S438P Mutant Forms Complexes with Other Components— To test whether the PS1 F411Y/S438P mutant forms a complex with other components, γ-secretase was immunopurified from yeast microsomes with the PS1 antibody against loop region (G1L3) (42) (Fig. 6A). NCT (mature and immature forms), Aph-1, and Pen2 were coimmunopurified with wild type PS1 or the F411Y/S438P mutant (Fig. 6, A and B, lanes 1 and 3). In the absence of NCT, Aph-1 and Pen2 were still copurified with wild type or F411Y/S438P PS1 (Fig. 6B, lanes 2 and 4). The recovery of NCT, Aph-1, and Pen2 was at a similar level between wild type and the F411Y/S438P mutant, suggesting that F411Y/S438P forms complexes with other subunits in the presence or absence of NCT.
FIGURE 6.
F411Y/S438P mutant forms γ-secretase complexes in yeast. A, microsomes from yeast transformants with PS1 wild type, NCT, Aph-1-HA, and FLAG-Pen2 were solubilized with buffer containing 1% CHAPSO. γ-Secretase complex was purified from the extracts using the antibody against PS1-loop region (G1L3) or rabbit preimmune serum, as indicated. Immunoprecipitates and the input fraction were analyzed by immunoblotting using specific antibodies. The input represents 25% of the microsomal extract. B, microsomes were prepared from yeast transformants expressing PS1 wild type or F411Y/S438P mutant, NCT, Aph-1-HA, and FLAG-Pen2 as indicated. IP, immunoprecipitation. γ-Secretase complexes were immunopurified by G1L3 antibody and analyzed by immunoblotting as A.
DISCUSSION
Presenilin is unique among proteases cleaving intramembrane domains because it requires regulatory subunits (NCT, Aph-1, and Pen2) for the activity. The functions of these subunits have yet to be understood. To address this problem, we have expressed the γ-secretase in yeast and screened PS1 mutants, which do not require other subunits for the protease activity. Fifteen mutants did not require NCT and exhibited wild type activity to cleave APP (C1–55)-Gal4p or NotchTM-Gal4p, and to endoproteolyze. Protease activity of these mutants was further confirmed by the endoproteolysis, the Aβ production from APP (C1–99), and the NICD production from Notch-1 (NΔE) in mouse fibroblasts. This finding led us to study the roles of trans-membrane helices of PS1, as discussed below.
Although high resolution structure of PS1 or other subunits is not established, structural considerations are important for understanding PS1 mutants active in the absence of NCT and thus γ-secretase itself. All of the mutants had S438P substitution together with a missense mutation, except that one had three replacements, V236M/F428Y/F411Y. The Ser438 is localized in the middle of the TM9, and its replacement to Pro residue may change the structure of the trans-membrane domain and induce flexibility, because Pro is a known α-helix breaker (44). Activation of the S438P mutant in the absence of NCT correlates with the proposed role of TM9 for the binding of the substrate (17) and the interaction with NCT (11–13). The Ser438 residue is conserved through PS1/PS2 of all species so far sequenced and locates very close to the motif (Pro-Ala-Leu-Pro-Ile-Ser, Pro433 to Ser438, in human residue numbers). It should also be noted that Pro-Ala-Leu (PAL motif, underlined above) is completely conserved among PS1, PS2, and SPP, and the mutation in the motif abolishes the γ-secretase activity (13) and NCT binding (11). Thus, the S438P mutation obviously affected the location of PAL motif. Recent cross-linking experiments indicated that the PAL motif and Ser438 exist very close to the catalytic Asp257 residue in TM6 (21). Tolia et al. (20) show that TM9 exhibits a highly flexible structure and propose that it may be involved in the transport of the substrates to the catalytic site. They suggest that TM6 (Asp257), TM7 (Asp385), TM9 (the PAL motif and Ser438), and the hydrophobic region between TM6 and TM7 (hydrophobic domain VII) form a catalytic cavity. We propose that the S438P mutation changed the structure of the recognition site, which facilitates the substrate entry into the catalytic site.
S438P mutant is partially active in the absence of NCT. However, additional single mutation was required for full protease activity. The secondary mutations are distributed through TM1, TM3, TM5, TM6, TM8, TM9, and loop regions. Among them, A246T and V252M may directly change the position of the catalytic Asp257 in TM6, and F411Y and I416F may change the structure of TM8, which interacts with Aph-1 and NCT. Immunoprecipitation experiments confirmed that F411Y/S438P mutant forms a complex with Aph-1 and Pen2 even in the absence of NCT. Pen2 binds to the pre-formed complex with PS1, Aph-1, and NCT and enhances the endoproteolysis upon maturation of γ-secretase (9). The interaction between the PS1 mutant and Pen2 may explain why the mutant was activated in the absence of NCT. Pen2 interacts with the TM4 of PS1 (14, 15). Thus, S438P and other mutations may change structural arrangement of TM4 and enhance the interaction between PS1 catalytic site and Pen2 in the absence of NCT.
Immunochemical results indicated that the mutant PS1, F411Y/S438P, became a mature form by endoproteolysis in yeast in the absence of NCT. The ratio of the endoproteolyzed fragments to PS1 increased with the mutant, suggesting that the mutant has higher endoproteolytic activity than wild type. In mammalian fibroblast cells, the mutant PS1 also endoproteolyzed in the absence of NCT. However, the production of the mature fragments was lower than that with NCT. This may be because the mutant is unstable or inactivated in fibroblast cells. It was shown that PS1 is degraded by proteasome in the absence of NCT (9).
When we observed the cleavage of the C1–99 fragment in vitro using solubilized yeast microsomes (38), we could detect neither Aβ nor AICD production by the F411Y/S438P mutant in the absence of NCT. The γ-secretase assay system required the CHAPSO solubilization of membranes (38). The activity of F411Y/S438P mutant in the absence of NCT may be sensitive to detergents. Because it is difficult to detect Aβ or AICD production in yeast whole cells (lysate or media), we further tested the C1–99 cleavage in mammalian whole cell assays.
Using the mouse embryonic fibroblast cells, we observed the Aβ and NICD production by the PS1 mutant, F411Y/S438P, in the absence of NCT, confirming its activity to cleave Gal4p-fused substrates. The processing of the physiological substrates (C1–99 or NΔE) in mammalian cells confirms that the mutant complex is active γ-secretase in the absence of NCT. Although F411Y/S438P mutant possessed the wild type activity to process Gal4-fused substrates in yeast, the Aβ or NICD production by F411Y/S438P mutant was low compared with the wild type PS1 with NCT in fibroblasts. When we tested the stability of F411Y/S438P by the cycloheximide chase experiment, we observed that the mature form of the mutant (CTF) is unstable (supplemental Fig. S2) compared with wild type PS1. This result corroborates the low activity of the mutant PS1. On the other hand, the subcellular localization of the mutant was tested by the sucrose gradient sedimentation. The wild type and the mutant PS1 were detected in the Golgi, the endoplasmic reticulum, and the plasma membrane fractions (supplemental Fig. S3). Distribution of PS1 CTF was similar between wild type and F411Y/S438P (supplemental Fig. S3), suggesting that the cleavage reaction may occur in similar organella. After extensive screening, we could isolate no PS1 mutant active in the absence of Pen2 or Aph-1. Because fully active PS1 without NCT required two amino acid replacements, those active without Pen2 or Aph-1 may also require multiple mutations. If that is the case, the screenings were not genetically saturated. Further screening is under way for PS1 mutants active without Aph-1 or Pen2 focusing mainly on the similarities between PS1 and its structural homologue SPP. The analyses of the FAD mutants of PS1 showed that G384A and L166P are loss of function like the active site mutant D385A. Using the yeast Gal4 assay, all FAD mutants showed reduced activity compared with the wild type. It is noteworthy that none of them had elevated protease activity. These results support the loss of function pathology of PS1 (7). As tested so far, all the PS1 FAD mutants required NCT for its activity. Using in vitro γ-secretase assay with yeast microsomes, we found that wild type PS1 produces Aβ40, Aβ42, and Aβ43 (38). It is of interest to analyze the abnormal production of toxic Aβ42 from the FAD mutant in yeast.
γ-Secretase is a prominent drug target for Alzheimer disease. However, its growing list of substrates (22), including developmentally indispensable Notch, have rendered inhibitor design challenging. Recently, one PS1 mutant, I437C in TM9, was found to block the Aβ production but not the Notch processing (20), suggesting that TM9 may contribute to substrate specificity. On the other hand, nonsteroidal anti-inflammatory drugs were found to modulate the protease function and block the Aβ42 production without affecting the Notch processing (45). We have developed two artificial substrates with APP or Notch in yeast. The yeast system has possibilities of drug search. It will be a great tool to screen mutations, genes, and drugs, which specifically block the Aβ production.
Supplementary Material
Acknowledgments
We thank Dr. Takeshi Iwatsubo and Dr. Taisuke Tomita (University of Tokyo) for PS1 antisera and Pen2 and Aph-1 clones; Dr. Takaomi C. Saido (RIKEN) for PS1 antisera; Dr. Raphael Kopan (Washington University) for the mNotch1 clone; Dr. Bart De Strooper (Vlaams Instituut voor Biotechnologie) for PS1/PS2 double knock-out cells; Dr. Philip C. Wong (The Johns Hopkins University) for NCT knock-out cells; and Dr. Philip James (University of Wisconsin) for the PJ-69-4A yeast strain. We also thank Dr. Randy Schekman (University of California, Berkeley), Dr. Tatsuya Maeda (University of Tokyo), and Dr. Taisuke Tomita for helpful discussions and technical suggestions. We also thank the members of our laboratory, especially Dr. Noboru Sasagawa, for encouragement and critical comments.
This work was supported by research grants from Human Frontier Science Program and the Ministry of Health, Labor, and Welfare, Japan (to S. I.), the Ministry of Education, Culture, Sports, Science, and Technology (to E. F. and S. Y.), The Uehara Memorial Foundation, and Takeda Chemical Science Foundation (to E. F.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1–S3, and Table S1.

Author's Choice—Final version full access..
Footnotes
The abbreviations used are: APP, amyloid precursor protein; Aβ, amyloid β peptide; AICD, APP intracellular domain; Aph-1, anterior pharynx-1; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid; CTF, carboxyl-terminal fragment; NCT, nicastrin; NICD, Notch intracellular domain; NTF, amino-terminal fragment; PS, presenilin; Pen2, presenilin enhancer 2; FAD, familial Alzheimer disease; PIPES, 1,4-piperazinediethanesulfonic acid; TM, transmembrane domain; siRNA, small interfering RNA.
References
- 1.Selkoe, D. J. (2001) Physiol. Rev. 81 741-766 [DOI] [PubMed] [Google Scholar]
- 2.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H., and Haass, C. (2003) Nat. Cell Biol. 5 486-488 [DOI] [PubMed] [Google Scholar]
- 3.Laudon, H., Hansson, E. M., Melén, K., Bergman, A., Farmery, M. R., Winblad, B., Lendahl, U., von Heijne, G., and Näslund, J. (2005) J. Biol. Chem. 280 35352-35360 [DOI] [PubMed] [Google Scholar]
- 4.Oh, Y. S., and Turner, R. J. (2005) Biochemistry 44 11821-11828 [DOI] [PubMed] [Google Scholar]
- 5.Spasic, D., Tolia, A., Dillen, K., Baert, V., De Strooper, B., Vrijens, S., and Annaert, W. (2006) J. Biol. Chem. 281 26569-26577 [DOI] [PubMed] [Google Scholar]
- 6.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T., and Selkoe, D. J. (1999) Nature 398 513-517 [DOI] [PubMed] [Google Scholar]
- 7.Shen, J., and Kelleher, R. J., III (2007) Proc. Natl. Acad. Sci. U. S. A. 104 403-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah, S., Lee, S. F., Tabuchi, K., Hao, Y. H., Yu, C., LaPlant, Q., Ball, H., Dann, C. E., 3rd, Südhof, T., and Yu, G. (2005) Cell 122 435-447 [DOI] [PubMed] [Google Scholar]
- 9.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G., and Iwatsubo, T. (2003) Nature 422 438-441 [DOI] [PubMed] [Google Scholar]
- 10.Niimura, M., Isoo, N., Takasugi, N., Tsuruoka, M., Ui-Tei, K., Saigo, K., Morohashi, Y., Tomita, T., and Iwatsubo, T. (2005) J. Biol. Chem. 280 12967-12975 [DOI] [PubMed] [Google Scholar]
- 11.Kaether, C., Capell, A., Edbauer, D., Winkler, E., Novak, B., Steiner, H., and Haass, C. (2004) EMBO J. 23 4738-4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergman, A., Laudon, H., Winblad, B., Lundkvist, J., and Näslund, J. (2004) J. Biol. Chem. 279 45564-45572 [DOI] [PubMed] [Google Scholar]
- 13.Wang, J., Beher, D., Nyborg, A. C., Shearman, M. S., Golde, T. E., and Goate, A. (2006) J. Neurochem. 96 218-227 [DOI] [PubMed] [Google Scholar]
- 14.Kim, S. H., and Sisodia, S. S. (2005) J. Biol. Chem. 280 41953-41966 [DOI] [PubMed] [Google Scholar]
- 15.Watanabe, N., Tomita, T., Sato, C., Kitamura, T., Morohashi, Y., and Iwatsubo, T. (2005) J. Biol. Chem. 280 41967-41967 [DOI] [PubMed] [Google Scholar]
- 16.Kornilova, A. Y., Bihel, F., Das, C., and Wolfe, M. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3230-3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Annaert, W. G., Esselens, C., Baert, V., Boeve, C., Snellings, G., Cupers, P., Craessaerts, K., and De Strooper, B. (2001) Neuron 32 579-589 [DOI] [PubMed] [Google Scholar]
- 18.Tolia, A., Chávez-Gutiérrez, L., and De Strooper, B. (2006) J. Biol. Chem. 281 27633-27642 [DOI] [PubMed] [Google Scholar]
- 19.Sato, C., Morohashi, Y., Tomita, T., and Iwatsubo, T. (2006) J. Neurosci. 26 12081-12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolia, A., Horré, K., and De Strooper, B. (2008) J. Biol. Chem. 283 19793-19803 [DOI] [PubMed] [Google Scholar]
- 21.Sato, C., Takagi, S., Tomita, T., and Iwatsubo, T. (2008) J. Neurosci. 28 6264-6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. Mol. Cell Biol. 5 499-504 [DOI] [PubMed] [Google Scholar]
- 23.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K., and Martoglio, B. (2002) Science 296 2215-2218 [DOI] [PubMed] [Google Scholar]
- 24.Martin, L., Fluhrer, R., Reiss, K., Kremmer, E., Saftig, P., and Haass, C. (2008) J. Biol. Chem. 283 1644-1652 [DOI] [PubMed] [Google Scholar]
- 25.Sato, T., Nyborg, A. C., Iwata, N., Diehl, T. S., Saido, T. C., Golde, T. E., and Wolfe, M. S. (2006) Biochemistry 45 8649-8656 [DOI] [PubMed] [Google Scholar]
- 26.Iben, L. G., Olson, R. E., Balanda, L. A., Jayachandra, S., Robertson, B. J., Hay, V., Corradi, J., Prasad, C. V., Zaczek, R., Albright, C. F., and Toyn, J. H. (2007) J. Biol. Chem. 282 36829-36836 [DOI] [PubMed] [Google Scholar]
- 27.Narayanan, S., Sato, T., and Wolfe, M. S. (2007) J. Biol. Chem. 282 20172-20179 [DOI] [PubMed] [Google Scholar]
- 28.Herreman, A., Hartmann, D., Annaert, W., Saftig, P., Craessaerts, K., Serneels, L., Umans, L., Schrijvers, V., Checler, F., Vanderstichele, H., Baekelandt, V., Dressel, R., Cupers, P., Huylebroeck, D., Zwijsen, A., Van Leuven, F., and De Strooper, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11872-11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herreman, A., Serneels, L., Annaert, W., Collen, D., Schoonjans, L., and De Strooper, B. (2000) Nat. Cell Biol. 2 461-462 [DOI] [PubMed] [Google Scholar]
- 30.Li, T., Ma, G., Cai, H., Price, D. L., and Wong, P. C. (2003) J. Neurosci. 23 3272-3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Y., Luo, W., Wang, H., Lin, P., Vetrivel, K. S., Liao, F., Li, F., Wong, P. C., Farquhar, M. G., Thinakaran, G., and Xu, H. (2005) J. Biol. Chem. 280 17020-17026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, C. A., III, Martinat, M. A., and Hyman, L. E. (1998) Nucleic Acids Res. 26 3577-3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mumberg, D., Müller, R., and Funk, M. (1995) Gene (Amst.) 156 119-122 [DOI] [PubMed] [Google Scholar]
- 34.James, P., Halladay, J., and Craig, E. A. (1996) Genetics 144 1425-1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clontech (2001) Yeast Protocols Handbook, Clontech, Mountain View, CA
- 36.Ono, Y., Torii, F., Ojima, K., Doi, N., Yoshioka, K., Kawabata, Y., Labeit, D., Labeit, S., Suzuki, K., Abe, K., Maeda, T., and Sorimachi, H. (2006) J. Biol. Chem. 281 18519-18531 [DOI] [PubMed] [Google Scholar]
- 37.Wuestehube, L. J., and Schekman, R. W. (1992) Methods Enzymol. 219 124-136 [DOI] [PubMed] [Google Scholar]
- 38.Yagishita, S., Futai, E., and Ishiura, S. (2008) Biochem. Biophys. Res. Commun. 377 141-145 [DOI] [PubMed] [Google Scholar]
- 39.Yagishita, S., Morishima-Kawashima, M., Ishiura, S., and Ihara, Y. (2008) J. Biol. Chem. 283 733-738 [DOI] [PubMed] [Google Scholar]
- 40.Yagishita, S., Morishima-Kawashima, M., Tanimura, Y., Ishiura, S., and Ihara, Y. (2006) Biochemistry 45 3952-3960 [DOI] [PubMed] [Google Scholar]
- 41.Schroeter, E. H., Kisslinger, J. A., and Kopan, R. (1998) Nature 393 382-386 [DOI] [PubMed] [Google Scholar]
- 42.Takasugi, N., Takahashi, Y., Morohashi, Y., Tomita, T., and Iwatsubo, T. (2002) J. Biol. Chem. 277 50198-50205 [DOI] [PubMed] [Google Scholar]
- 43.Tomita, T., Maruyama, K., Saido, T. C., Kume, H., Shinozaki, K., Tokuhiro, S., Capell, A., Walter, J., Grunberg, J., Haass, C., Iwatsubo, T., and Obata, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2025-2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consler, T. G., Tsolas, O., and Kaback, H. R. (1991) Biochemistry 30 1291-1298 [DOI] [PubMed] [Google Scholar]
- 45.Weggen, S., Eriksen, J. L., Das, P., Sagi, S. A., Wang, R., Pietrzik, C. U., Findlay, K. A., Smith, T. E., Murphy, M. P., Bulter, T., Kang, D. E., Marquez-Sterling, N., Golde, T. E., and Koo, E. H. (2001) Nature 414 212-216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.