Abstract
Endothelial apoptosis is a pivotal process for angiogenesis during embryogenesis as well as postnatal life. By using a retrovirus-mediated signal sequence trap method, we identified a previously undescribed gene, termed ARIA (apoptosis regulator through modulating IAP expression), which regulates endothelial apoptosis and angiogenesis. ARIA was expressed in blood vessels during mouse embryogenesis, as well as in endothelial cells both in vitro and in vivo. ARIA is a unique protein with no homology to previously reported conserved domain structures. Knockdown of ARIA in HUVECs by using small interfering RNA significantly reduced endothelial apoptosis without affecting either cell migration or proliferation. ARIA knockdown significantly increased inhibitor of apoptosis (cIAP)-1 and cIAP-2 protein expression, although their mRNA expression was not changed. Simultaneous knockdown of cIAP-1 and cIAP-2 abolished the antiapoptotic effect of ARIA knockdown. Using yeast 2-hybrid screening, we identified the interaction of ARIA with 20S proteasome subunit α-7. Thereafter, we found that cIAP-1 and cIAP-2 were degraded by proteasomes in endothelial cells under normal condition. Overexpression of ARIA significantly reduced cIAP-1 expression, and this reduction was abolished by proteasomal inhibition in BAECs. Also, knockdown of ARIA demonstrated an effect similar to proteasomal inhibition with respect to not only expression but also subcellular localization of cIAP-1 and cIAP-2. In vivo angiogenesis studied by Matrigel-plug assay, mouse ischemic retinopathy model, and tumor xenograft model was significantly enhanced by ARIA knockdown. Together, our data indicate that ARIA is a unique factor regulating endothelial apoptosis, as well as angiogenesis, presumably through modulating proteasomal degradation of cIAP-1 and cIAP-2 in endothelial cells.
Angiogenesis is the process of forming new blood vessels through sprouting and budding of new capillaries from existing blood vessels. Endothelial cells constitute the inner layer of blood vessels and have critical roles in angiogenesis under both physiological and pathological conditions. Because of the central role of angiogenesis in ischemic cardiovascular diseases, as well as in cancer, modulating this process is a promising approach to treat these diseases. In fact, enhancing angiogenesis by administration of growth factors or cell transplantation to treat ischemic diseases and reducing angiogenesis by inhibiting VEGF to treat cancer have been clinically used and demonstrated considerably beneficial effects (1–4). However, use of these therapies is not yet satisfactory, and improvement is certainly needed. To develop better therapeutic angiogenesis, it is crucial to understand its detailed molecular mechanism, including unknown factors regulating endothelial cell function.
Membrane proteins, as well as secreted proteins expressed in endothelial cells, are known to have specific and critical roles in the regulation of endothelial function and angiogenesis (5–7). To identify novel factors regulating endothelial function and angiogenesis, we performed a signal sequence trap screening that specifically traps genes encoding secreted and membrane proteins. Here, we characterize a previously undescribed gene, named ARIA (apoptosis regulator through modulating IAP expression). ARIA is expressed in endothelial cells both in vitro and in vivo, as well as in blood vessels, during mouse embryogenesis. Knockdown of ARIA expression in human umbilical vein endothelial cells (HUVECs) significantly reduced apoptosis by increasing inhibitor of apoptosis (cIAP)-1 and cIAP-2 protein expression. In vitro and in vivo studies indicate a significant role for this factor in angiogenesis.
Results
Isolation of ARIA.
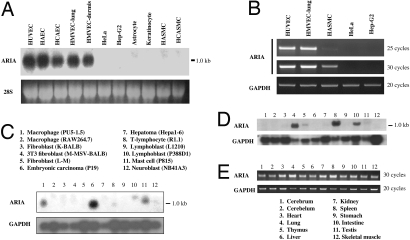
To isolate novel factors regulating endothelial cell function and angiogenesis, we have employed a signal sequence trap using a cDNA library prepared from human microvascular endothelial cells. One gene we have isolated demonstrated significant expression in endothelial cells, whereas no expression was observed in nonendothelial cells (Fig. 1A). We named this gene ARIA and further analyzed its function in endothelial cells. ARIA was also expressed in vascular smooth muscle cells, as well as in hematopoietic cells such as macrophages, lymphocytes, and mast cells, although those expression levels appeared to be much lower than in endothelial cells (Fig. 1 B and C). In adult mouse tissues, ARIA was expressed in all tissues examined, and the highest expression was observed in lung and spleen (Fig. 1 D and E). In situ hybridization of ARIA in embryonic day 9.5 (e9.5d) mouse embryo demonstrated its expression in blood vessels (Fig. 2A and B). ARIA was expressed as early as e7.5d, and its expression was maintained at least until e15.5d (Fig. 2C). These results suggest that ARIA might be a previously undescribed factor regulating endothelial cell function and angiogenesis.
Fig. 1.
Messenger RNA expression of ARIA. (A) Northern blot analysis of ARIA in primary cultured human endothelial cells and other cultured human cells. Cells examined were HUVEC, human aortic endothelial cells (HAEC), human coronary artery endothelial cells (HCAEC), human lung microvascular endothelial cells (HMVEC-lung), human dermal microvascular endothelial cells (HMVEC-dermis), HeLa, human hepatoma cells (Hep-G2), human astrocytes, human keratinocytes, human aortic smooth muscle cells (HASMC), and human coronary artery smooth muscle cells (HCASMC). (B) RT-PCR analysis of ARIA. Little expression of ARIA was observed in vascular smooth muscle cells, but not in HeLa or Hep-G2 cells. (C) Northern blot analysis of ARIA in mouse cell line MTN blot (BD Clontech). ARIA was expressed in some of hematopoietic cells. (D) Northern blot analysis of ARIA in mouse tissues. (E) RT-PCR analysis of ARIA in mouse tissues.
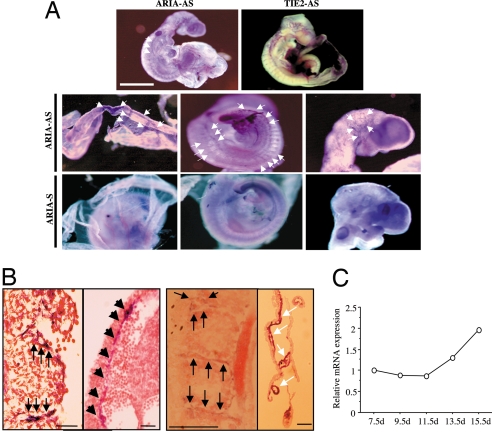
Fig. 2.
Expression of ARIA during mouse embryogenesis. (A) Whole-mount in situ hybridization of ARIA in e9.5d mouse embryo. ARIA was expressed in blood vessels during embryogenesis as indicated by arrows (ARIA-AS). Negative control using sense cRNA probe (ARIA-S) did not show significant signals. In situ hybridization of TIE-2, expressed almost exclusively in endothelial cells and early hematopoietic cells, was performed for a positive control of the blood vessel expression. (B) Sections of embryo from the whole-mount in situ hybridization. ARIA expression was observed in intersomitic arteries, dorsal aorta (black arrows), and blood vessels on the yolk sac (white arrows). (Scale bars, 100 μm.) (C) Relative expression of ARIA in the whole embryos at indicated embryonic day.
Expression of ARIA in Vitro and in Vivo.
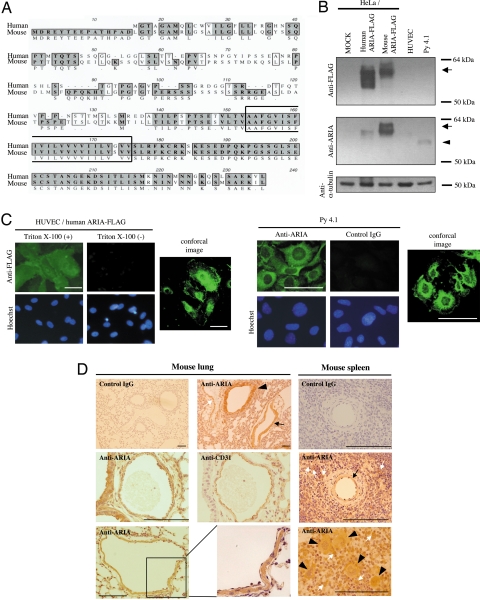
Full-length human and mouse ARIA cDNAs were isolated as described in Materials and Methods. The sequences of these genes have been submitted to the GenBank database under accession nos. EU025066 (human ARIA) and EU025067 (mouse ARIA). The amino acid sequences of both human and mouse ARIA contained a putative transmembrane domain (Fig. 3A). Overall amino acid sequence homology between human and mouse ARIA was ≈61%. However, when focusing on the C-terminal region, including the putative transmembrane domain, their homology was ≈91%, suggesting that this region may have an important role in ARIA function. When expressed in HeLa cells, recombinant proteins tagged with FLAG of both human and mouse ARIA migrated at ≈60 kDa, which is significantly larger than the predicted molecular mass (≈24 kDa), suggesting that ARIA undergoes posttranslational modification (Fig. 3B). We generated anti-mouse ARIA antibody by using a mixture of 3 epitopes of mouse ARIA. This antibody successfully detected endogenous ARIA in mouse endothelial cells (py4.1), but did not cross-react well with human ARIA, and failed to detect it in HUVECs (Fig. 3B). Immunocytochemistry using anti-FLAG antibody demonstrated that human ARIA was expressed in cytosol, as well as on plasma membrane (Fig. 3C). A FLAG epitope tag was added at the C terminus for the detection of recombinant protein. Because no signal was observed without permeabilization treatment with Triton X-100, the C terminus of membrane-bound ARIA is likely located in the intracellular region (Fig. 3C). Endogenous mouse ARIA detected by the anti-ARIA antibody was also localized both in cytosol and on plasma membrane in py4.1 mouse endothelial cells (Fig. 3C).
Fig. 3.
Protein expression of ARIA. (A) Amino acid sequences of human and mouse ARIA. Putative membrane domain is surrounded by a solid line. The same amino acids between human and mouse ARIA are highlighted, and similar amino acids are indicated by open boxes. (B) Human and mouse ARIA tagged with FLAG was expressed in HeLa cells and followed by immunoblotting using anti-FLAG M2 antibody (arrow). Recombinant as well as endogenous mouse ARIA was detected by anti-mouse ARIA antibody (arrow and arrowhead, respectively). (C) Human ARIA tagged with FLAG at its C terminus was expressed in HUVECs as shown by immunocytochemistry using anti-FLAG M2 antibody. Expression of human ARIA-FLAG was detected in cytosol as well as on plasma membrane when cells were permeabilized with 0.1% Triton X-100. Endogenous mouse ARIA detected by the anti-ARIA antibody was also localized both in cytosol and on plasma membrane in py4.1 cells. (Scale bars, 100 μm.) (D) Expression of ARIA in mouse lung and spleen. ARIA expression was detected in bronchial epithelial cells (arrowheads), artery (black arrow), and vein (white arrow) in lung. In blood vessels, ARIA was expressed in both endothelial and vascular smooth muscle cells. ARIA was also expressed in blood vessels in spleen (black arrow). A subset of lymphocytes (white arrows) as well as macrophages (arrowhead) expressed ARIA in spleen. (Scale bars, 100 μm.)
Immunohistochemistry of ARIA in mouse lung and spleen demonstrated its expression in endothelial cells, as well as in vascular smooth muscle cells, bronchial epithelial cells, a subset of lymphocytes, and macrophages (Fig. 3D). These in vivo expression profiles of ARIA were consistent with the in vitro expression analysis.
ARIA Regulates Endothelial Apoptosis.
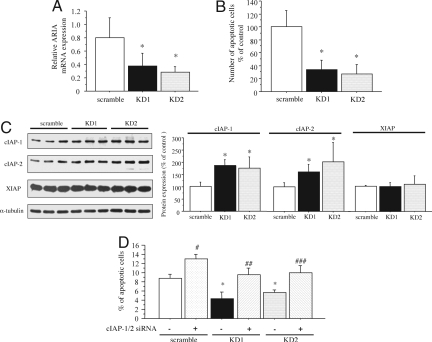
To investigate ARIA function in endothelial cells, we prepared 2 independent siRNAs to knock down ARIA expression. Negative control siRNA (scramble siRNA) was used as a control. We have confirmed effective transfection of siRNA in HUVECs by electroporation, as well as by using RNAiMAX reagent (Fig. S1A). Both ARIA siRNAs (KD1 and KD2) effectively knocked down ARIA mRNA expression in HUVECs by electroporation (Fig. 4A) and by using RNAiMAX reagent (Fig. S1B). When apoptosis was induced by serum and growth factor depletion, ARIA knockdown resulted in significant reduction of endothelial apoptosis as compared with cells transfected with the scramble siRNA (Fig. 4B). In contrast, ARIA knockdown did not affect either endothelial cell migration or proliferation (Fig. S2), suggesting that ARIA is closely involved in the regulation of endothelial apoptosis.
Fig. 4.
ARIA-knockdown reduced endothelial cell apoptosis by modulating cIAP-1 and cIAP-2 expression. (A) ARIA expression was successfully knocked down by the 2 independent siRNA (KD1 and KD2). *, P < 0.05 versus cells transfected with the negative control scramble siRNA (n = 5 each). (B) ARIA-knockdown significantly reduced HUVEC apoptosis induced by serum and growth factor depletion. Tunel-positive cells were counted in an equal area and corrected by the total cell number. *, P < 0.01 versus cells transfected with the scramble siRNA (n = 4 each). (C) ARIA-knockdown increased cIAP-1 and cIAP-2 but not XIAP expression in HUVEC. *, P < 0.05 versus cells transfected with the scramble siRNA (n = 7 each). (D) Simultaneous knockdown of cIAP-1 and cIAP-2 completely abolished the anti-apoptotic effect of ARIA-knockdown. *, #, P < 0.01 versus cells transfected with only the scramble siRNA. ##, P < 0.01 versus cells transfected with only KD1. ###, P < 0.01 versus cells transfected with only KD2. ##, ###, NS versus cells transfected with only the scramble siRNA (n = 7 each).
Also, analysis of the expressional regulation of ARIA revealed that some cytokines such as TNF-α and TGF-β decreased ARIA expression in endothelial cells (Fig. S3).
ARIA Regulates Endothelial Apoptosis by Modulating cIAP-1 and cIAP-2 Expression.
To elucidate the molecular mechanism responsible for the antiapoptotic effect of ARIA knockdown, signals and molecules regulating cell apoptosis were analyzed by immunoblotting. Phosphorylation of MAPK families, as well as Akt, was not affected by ARIA knockdown (Fig. S4A). We also observed that reactive oxygen species production assessed by dichlorodihydrofluorescein (DCF) fluorescence was not affected by the ARIA knockdown (Fig. S4B). Finally, we identified that cIAP-1 (Birc2) and cIAP-2 (Birc3) protein expression was significantly increased in HUVECs as a result of ARIA knockdown (Fig. 4C). In contrast, X-linked IAP (XIAP) was not increased by ARIA knockdown (Fig. 4C). Simultaneous knockdown of cIAP-1 and cIAP-2 abrogated the antiapoptotic effect of ARIA knockdown (Fig. 4D). Knockdown of cIAP-1 and cIAP-2 was confirmed at the protein level, as well as at the mRNA level (Fig. S5). These results indicate that ARIA regulates endothelial cell survival through modulating cIAP-1 and cIAP-2 expression.
Despite their increased protein expression, mRNA level of both cIAP-1 and cIAP-2 was not increased by ARIA knockdown (Fig. S5C). These results suggest that ARIA probably modulates cIAP-1 and cIAP-2 expression at the protein level in endothelial cells.
ARIA Modulates Proteasomal Degradation of cIAP-1 and cIAP-2.
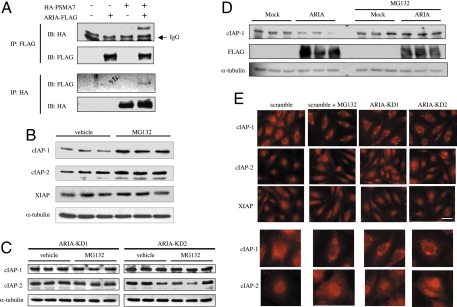
To identify the binding partner of ARIA in HUVECs, we have performed yeast 2-hybrid screening using the highly conserved C-terminal region of human ARIA as bait. One positive clone encodes 70 aa of C-terminal region of 20S proteasome subunit α (PSMA)-7. PSMA-7 is one of the subunits constituting cylinder-shaped 20S core proteasome and is located at the outer rings flanking an internal pair of β rings (8). We confirmed their interaction by coprecipitation of HA-tagged PSMA-7 and FLAG-tagged ARIA expressed in bovine aortic endothelial cells (BAECs) (Fig. 5A).
Fig. 5.
ARIA modulates proteasomal degradation of cIAP-1 and cIAP-2. (A) HA-tagged PSMA-7 and/or FLAG-tagged ARIA were expressed in BAECs. HA-PSMA7 and ARIA-FLAG were immunoprecipitated by anti-HA antibody and anti-FLAG M2 antibody, respectively. Coprecipitation of HA-PSMA7 and ARIA-FLAG was observed. (B) Proteasome inhibition with MG132 significantly increased cIAP-1 and cIAP-2, but not XIAP, expression in HUVECs. Similar results were obtained by 3 independent experiments. (C) However, when ARIA was knocked down, proteasome inhibition failed to further increase cIAP-1 and cIAP-2 expression. Similar results were obtained in 2 independent experiments. (D) Overexpression of human ARIA significantly reduced cIAP-1 expression in BAECs. Proteasome inhibition with MG132 abrogated this effect of ARIA overexpression on cIAP-1 expression. Similar results were obtained in 2 independent experiments. (E) Expression and subcellular localization of cIAP-1, cIAP-2, and XIAP in HUVECs. An effect of ARIA knockdown similar to proteasomal inhibition was observed with respect to expression as well as subcellular localization of cIAP-1 and cIAP-2. (Scale bar, 100 μm.) Similar results were obtained in 3 independent experiments.
Although it has been reported that IAPs undergo proteasomal degradation on apoptotic stimuli in thymocytes (9), it is unclear whether they are also degraded by proteasomes in endothelial cells, especially under normal conditions. Interaction of ARIA with the proteasome subunit urged us to investigate its function in the proteasomal degradation of cIAP-1 and cIAP-2. We first explored whether cIAP-1 and cIAP-2 are degraded by proteasomes in endothelial cells under normal conditions. Inhibition of proteasomes by MG132 significantly increased cIAP-1 and cIAP-2, but not XIAP, expression in HUVECs, indicating that cIAP-1 and cIAP-2 are indeed degraded by proteasomes in endothelial cells under normal conditions (Fig. 5B).
In contrast, treatment with MG132 failed to further increase cIAP-1 and cIAP-2 expression in HUVECs when ARIA was knocked down (Fig. 5C). These results suggest that proteasomal degradation of cIAP-1 and cIAP-2 was reduced by ARIA knockdown. Also, overexpression of ARIA significantly reduced cIAP-1 expression in BAECs (Fig. 5D). Notably, this effect of ARIA overexpression was abolished by proteasome inhibition, indicating that ARIA positively regulates the proteasomal degradation of cIAP-1 in endothelial cells (Fig. 5D). Because our antibody did not recognize bovine cIAP-2, we could not assess cIAP-2 expression in BAECs.
We then investigated expression and subcellular localization of cIAP-1, cIAP-2, and XIAP in HUVECs by immunocytochemistry. Interestingly, they demonstrated distinct subcellular localization; cIAP-1 was in cytosol, cIAP-2 was largely in nucleus, and XIAP was in both cytosol and nucleus in the control HUVECs (Fig. 5E). When proteasomal degradation was inhibited by MG132, cIAP-1 accumulated in the perinuclear region with the appearance of small structures, whereas cytosolic expression of cIAP-2 was evenly enhanced (Fig. 5E). Expression and subcellular localization of XIAP were not significantly affected by MG132 treatment. Of note, expression, as well as subcellular localization of cIAP-1 and cIAP-2 in ARIA-knocked-down HUVECs were very similar to those in the control cells treated with MG132 (Fig. 5E).
To examine whether ARIA is involved in the proteasomal degradation specifically for cIAP-1 and cIAP-2, we investigated the effect of ARIA knockdown on the expression of other proteins targeted by the proteasomal degradation. As shown in Fig. S4, ARIA knockdown did not affect the protein expression of Bcl-2 and p53, well-known targets for proteasomal degradation (10). Also, ARIA knockdown did not affect the protein expression of survivin, IκB-α, or p21WAF1/CIP, all of which are targets for proteasomal degradation (Fig. S6) (10, 11). Survivin is another member of the IAP family of proteins, whose expression is regulated by proteasomal degradation in a cell cycle-dependent manner (12). Therefore, ARIA seems to modulate proteasomal degradation specifically for cIAP-1 and cIAP-2.
Taking these data together, ARIA positively regulates proteasomal degradation of cIAP-1 and cIAP-2, and, thus, ARIA knockdown enhances endothelial cell survival by increasing cIAP-1 and cIAP-2 expression.
ARIA Regulates Angiogenesis both in Vitro and in Vivo.
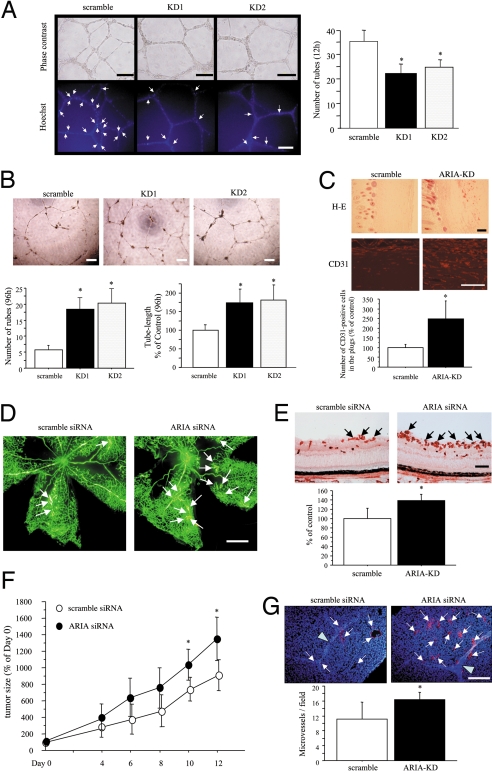
Because endothelial apoptosis is closely involved in angiogenesis (13. 14), we investigated the ARIA function in angiogenesis. Somewhat unexpectedly, ARIA knockdown caused a small reduction of tube-like structures of HUVECs on Matrigel at an early time point (12 h) (Fig. 6A). Fewer apoptotic endothelial cells were observed when ARIA was knocked down (Fig. 6A). Nevertheless, 96 h after plating, significantly greater tube-like structures were preserved in ARIA-knocked-down HUVECs (Fig. 6B).
Fig. 6.
ARIA regulates angiogenesis in vitro and in vivo. (A) ARIA knockdown reduced HUVEC tube formation on Matrigel at an early time point (12 h). *, P < 0.01 versus cells transfected with the scramble siRNA. Similar results were obtained in 3 independent experiments. Apoptotic cells on Matrigel were detected by Hoechst 33342 nuclear staining. Fewer apoptotic cells showing bright and condensed nuclei indicated by arrows were observed in ARIA-knocked-down HUVECs. (B) However, tube-structures were well preserved in ARIA-knocked-down HUVECs at a later time point (96 h) as compared with the control cells. *, P < 0.01 versus cells transfected with the scramble siRNA. Similar results were obtained in 2 independent experiments. (Scale bars, 200 μm.) (C) H&E staining as well as CD31 immunostaining of sections demonstrated significantly enhanced angiogenesis in the Matrigel plugs containing ARIA siRNA (ARIA-KD). (Scale bars, 100 μm.) CD31-positive endothelial cells were counted in an equal area. *, P < 0.01 versus the scramble control (n = 5 each). (D) Ischemic retinopathy was induced as described in Materials and Methods. Neovascularization (white arrows) was enhanced in retinas injected with ARIA siRNA. (Scale bar, 0.5 mm.) (E) Neovessels at the surface of retinas were detected by lectin staining (black arrows). The area of neovessels was significantly greater in the retinas injected with ARIA siRNA than in control. *, P < 0.05 versus the scramble control (n = 5 each). (Scale bar, 100 μm.) (F) Either scramble or ARIA siRNA was injected intratumorally at days 0, 4, and 8. The relative tumor volume (percentage of tumor volume on day 0) is presented in the graph. *, P < 0.05 versus tumors injected with the scramble siRNA (n = 6 each). (G) The number of CD31-positive microvessels (arrows) was significantly greater in the tumors injected with ARIA siRNA than in control. Arrowheads indicate the scars made by needle at the time of siRNA injection. *, P < 0.05 versus the scramble control (n = 6 each). (Scale bar, 100 μm.)
We then investigated the effect of ARIA knockdown in in vivo angiogenesis. Knockdown of ARIA expression by the mouse ARIA siRNA was validated in py4.1 mouse endothelial cells (Fig. S7A). A pair of Matrigel plugs containing either scramble or ARIA siRNA were implanted into mice at the bilateral flank, as shown in Fig. S8, followed by extraction at day 8. Extracted Matrigel plugs containing scramble siRNA were mostly pale, whereas plugs containing ARIA siRNA appeared yellowish (Fig. S8). ARIA expression in CD31-positive endothelial cells was significantly knocked down in the plugs containing the ARIA siRNA (Fig. S7B). H&E staining, as well as CD31 immunostaining of sections, revealed significantly enhanced angiogenesis by ARIA knockdown (Fig. 6C).
We further investigated the in vivo effect of ARIA knockdown on angiogenesis by using the mouse ischemic retinopathy model. Neovascularization in the retinas of ischemic retinopathy was enhanced, and the formation of neovessels detected by lectin staining in the surface of retinas was significantly increased by intravitreal injection of ARIA siRNA (Fig. 6 D and E). Also, fewer apoptotic endothelial cells were observed in the retinas injected with ARIA siRNA as compared with the scramble control (Fig. S9A). ARIA expression was considerably knocked down in the retinas injected with ARIA siRNA (Fig. S9B). These findings strongly suggest that ARIA knockdown enhances retinal angiogenesis by reducing endothelial apoptosis as presumed through the in vitro studies.
We further examined the role of ARIA in angiogenesis in vivo by using the tumor xenograft model. Intratumoral injection of ARIA siRNA significantly enhanced tumor growth as compared with the scramble control (Fig. 6F). ARIA expression was considerably knocked down in the tumor xenografts injected with ARIA siRNA (Fig. S9C). The number of microvessels was also significantly increased in the tumors injected with ARIA siRNA (Fig. 6G).
Together, our data indicate the potentially important role of ARIA in angiogenesis, presumably through modulating endothelial apoptosis.
Discussion
Angiogenesis is critically involved in the pathophysiology of ischemic cardiovascular diseases, as well as tumorigenesis; thus, regulating angiogenesis is a promising approach to treat such diseases. In this study, we described a molecular identification and characterization of a previously undescribed gene, termed ARIA. ARIA is expressed preferentially in endothelial cells, and knockdown of ARIA reduces endothelial apoptosis in vitro and enhances angiogenesis in vivo.
Members of the IAP family appear to act as “guardians” of the cell death machinery by directly binding to and neutralizing caspases, as well as by modulating the death signaling through TNF receptors (15–17). On apoptotic stimuli, IAPs autoubiquitylate themselves, and are rapidly degraded by proteasomes in thymocytes (9, 18). In the present study, we demonstrated a significant role of proteasomal degradation to determine the basal expression level of cIAP-1 and cIAP-2 in endothelial cells without apoptotic stimuli. Also, we revealed that knockdown of ARIA increased cIAP-1 and cIAP-2 expression, presumably through reducing their proteasomal degradation. Therefore, our data define not only a previously undescribed factor regulating endothelial apoptosis but also a previously undescribed pathway regulating cIAP-1 and cIAP-2 expression in endothelial cells.
We presume that the interaction of ARIA with PSMA-7 might have an important role in the regulation of cIAP-1 and cIAP-2 proteasomal degradation. Indeed, it has been reported that interaction of MDM2 with PSMA-3, as well as interaction of Tax with PSMA-4, has crucial roles in the proteasomal degradation of retinoblastoma protein and NF-κB, respectively (19, 20). Interaction with PSMA-3 has also been reported to be important for the proteasomal degradation of p21WAF1/CIP (21).
Endothelial apoptosis is known to have an important role in angiogenesis (14, 22–24). Endothelial apoptosis is observed in various physiological and pathological conditions such as wound healing, scar formation, atherosclerosis, and diabetic eye disease in the adult, as well as in developing capillaries during embryogenesis (22). In the present study, we demonstrated that knockdown of ARIA enhanced endothelial cell survival in vitro and angiogenesis in vivo, although it reduced in vitro tube formation of HUVECs at an early time point. These results suggest that endothelial apoptosis might have a biphasic function in angiogenesis. Endothelial apoptosis is probably essential for the initial lumen formation, but once the capillary network has been formed, inhibition of endothelial apoptosis might contribute to maintain it and prevent the involution of the newly formed network.
ARIA function in nonendothelial cells still remains unclear. Because IAPs have crucial roles in the cell survival and death in wide variety of cells, including hematopoietic cells (25–27), ARIA may also regulate the apoptosis of hematopoietic cells.
We demonstrated the significant proangiogenic effect of ARIA knockdown by the in vivo experiments such as Matrigel-plug assay, the ischemic retinopathy model, and the tumor xenograft model. Because these present studies are not performed by gene knockout using the ES cell system, but by gene knockdown using siRNA methods, we should cautiously conclude that ARIA has the in vivo function as an apoptosis-stimulating and antiangiogenic gene. Further studies using the knockout mice will be required to really define the proapoptotic and antiangiogenic function of ARIA in vivo.
In the ischemic animal models, administration of growth factors induces new vessel formation at early time point, but many of them regress as growth factors diminish over time (28). Thus, enhancing endothelial cell survival is very important to maintain growth factor-induced angiogenesis. Because ARIA appears to be expressed preferentially in endothelial cells, inhibition of ARIA might be a feasible approach for enhancing, as well as supporting, growth factor-induced angiogenesis, and ARIA might be an attractive new target for pharmacotherapeutic agents to treat ischemic diseases.
Materials and Methods
Cloning of ARIA.
Signal sequence trapping was performed as previously described (29). Nucleotide sequencing of one clone (ARIA) demonstrated no significant homology to genes identified before. The GenBank database was searched by using this partial nucleotide sequence to obtain human and mouse ARIA contiguous sequences. The nucleotide sequence of the 5′ end of human ARIA cDNA was determined by 5′-rapid amplification of cDNA ends. Last, full-length cDNA of human and mouse ARIA was obtained by RT-PCR, and the nucleotide sequence of both strands of cDNA was analyzed at least twice.
In Situ Hybridization.
Whole-mount in situ hybridization was performed using digoxigenin-labeled cRNA as previously described (30). After all procedures, embryos were embedded in OCT, snap-frozen, and sectioned. Sections were counterstained with eosin.
Western Blot Analysis.
Cell lysates were prepared in RIPA buffer, and then Western blot analysis was performed as previously described (31). For proteasomal inhibition, cells were treated with 10 μM MG132 (Calbiochem) for 6–8 h in the growth medium.
Mouse Ischemic Retinopathy Model.
Ischemic retinopathy in neonatal mice was produced in C57BL/6J mice as previously described (32–34). Briefly, postnatal day (P)7 mice were exposed to 75 ± 2% oxygen for 5 days (P7–P12) and then placed in room air for 5 days (P12–P17); 1 μg of scramble or ARIA siRNA was injected intravitreally immediately after placing in the room air. Methods are described in more detail in SI Materials and Methods.
Tumor Model.
B16 melanoma cells were s.c. inoculated in mice followed by the intratumoral siRNA injection. Methods are described in more detail in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Toshio Kitamura (Tokyo University) for providing the pMX-SST vector, Dr. Iichiro Shimomura (Osaka University) for providing a detailed protocol of signal sequence trapping, Dr. Garry Nolan (Stanford University) for providing the Phoenix-Eco packaging cells, and Dr. Hirokazu Yokoi and Yoshinori Tsubakimoto for excellent technical support. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) Grants 18790507 and 20590885, a Sakakibara Memorial Research Grant from the Japan Research Promotion Society for Cardiovascular Diseases, the Takeda Science Foundation, and the Japan Heart Foundation/Novartis Research Award on Molecular and Cellular Cardiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU025066 (human ARIA) and EU025067 (mouse ARIA)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0806780106/DCSupplemental.
References
- 1.Tateishi-Yuyama E, et al. Therapeutic angiogenesis for patients with limb ischaemia by autologous transplantation of bone-marrow cells: A pilot study and a randomised controlled trial. Lancet. 2002;360:427–435. doi: 10.1016/S0140-6736(02)09670-8. [DOI] [PubMed] [Google Scholar]
- 2.Kastrup J. Therapeutic angiogenesis in ischemic heart disease: Gene or recombinant vascular growth factor protein therapy? Curr Gene Ther. 2003;3:197–206. doi: 10.2174/1566523034578366. [DOI] [PubMed] [Google Scholar]
- 3.Lei Y, Haider H, Shujia J, Sim ES. Therapeutic angiogenesis. Devising new strategies based on past experiences. Basic Res Cardiol. 2004;99:121–132. doi: 10.1007/s00395-004-0447-x. [DOI] [PubMed] [Google Scholar]
- 4.Verheul HM, Pinedo HM. Vascular endothelial growth factor and its inhibitors. Drugs Today. 2003;39:81–93. [PubMed] [Google Scholar]
- 5.DeLisser HM, Newman PJ, Albelda SM. Platelet endothelial cell adhesion molecule (CD31) Curr Top Microbiol Immunol. 1993;184:37–45. doi: 10.1007/978-3-642-78253-4_3. [DOI] [PubMed] [Google Scholar]
- 6.Peters KG, et al. Functional significance of Tie2 signaling in the adult vasculature. Recent Prog Horm Res. 2004;59:51–71. doi: 10.1210/rp.59.1.51. [DOI] [PubMed] [Google Scholar]
- 7.Shibuya M. Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J Biochem Mol Biol. 2006;39:469–478. doi: 10.5483/bmbrep.2006.39.5.469. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K. Molecular biology of the proteasome. Biochem Biophys Res Commun. 1998;247:537–541. doi: 10.1006/bbrc.1998.8617. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 10.Thompson SJ, Loftus LT, Ashley MD, Meller R. Ubiquitin-proteasome system as a modulator of cell fate. Curr Opin Pharmacol. 2008;8:90–95. doi: 10.1016/j.coph.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond? Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Tenev T, Martins LM, Downward J, Lemoine NR. The ubiquitin-proteasome pathway regulates survivin degradation in a cell cycle-dependent manner. J Cell Sci. 2000;23:4363–4371. doi: 10.1242/jcs.113.23.4363. [DOI] [PubMed] [Google Scholar]
- 13.Segura I, et al. Inhibition of programmed cell death impairs in vitro vascular-like structure formation and reduces in vivo angiogenesis. FASEB J. 2002;16:833–841. doi: 10.1096/fj.01-0819com. [DOI] [PubMed] [Google Scholar]
- 14.Pollman MJ, Naumovski L, Gibbons GH. Endothelial cell apoptosis in capillary network remodeling. J Cell Physiol. 1999;178:359–370. doi: 10.1002/(SICI)1097-4652(199903)178:3<359::AID-JCP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 15.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 16.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ditzel M, Meier P. IAP degradation: Decisive blow or altruistic sacrifice? Trends Cell Biol. 2002;12:449–452. doi: 10.1016/s0962-8924(02)02366-8. [DOI] [PubMed] [Google Scholar]
- 18.Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- 19.Sdek P, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Rousset R, Desbois C, Bantignies F, Jalinot P. Effects on NF-kappa B1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature. 1996;381:328–331. doi: 10.1038/381328a0. [DOI] [PubMed] [Google Scholar]
- 21.Touitou R, et al. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duval H, Harris M, Li J, Johnson N, Print C. New insights into the function and regulation of endothelial cell apoptosis. Angiogenesis. 2003;6:171–183. doi: 10.1023/B:AGEN.0000021390.09275.bc. [DOI] [PubMed] [Google Scholar]
- 23.Choi ME, Ballermann BJ. Inhibition of capillary morphogenesis and associated apoptosis by dominant negative mutant transforming growth factor-beta receptors. J Biol Chem. 1995;270:21144–21150. doi: 10.1074/jbc.270.36.21144. [DOI] [PubMed] [Google Scholar]
- 24.Weihua Z, Tsan R, Schroit AJ, Fidler IJ. Apoptotic cells initiate endothelial cell sprouting via electrostatic signaling. Cancer Res. 2005;65:11529–11535. doi: 10.1158/0008-5472.CAN-05-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Graaf AO, de Witte T, Jansen JH. Inhibitor of apoptosis proteins: New therapeutic targets in hematological cancer? Leukemia. 2004;18:1751–1759. doi: 10.1038/sj.leu.2403493. [DOI] [PubMed] [Google Scholar]
- 26.Wrzesien-Kus A, Smolewski P, Sobczak-Pluta A, Wierzbowska A, Robak T. The inhibitor of apoptosis protein family and its antagonists in acute leukemias. Apoptosis. 2004;9:705–715. doi: 10.1023/B:APPT.0000045788.61012.b2. [DOI] [PubMed] [Google Scholar]
- 27.Akyurek N, Ren Y, Rassidakis GZ, Schlette EJ, Medeiros LJ. Expression of inhibitor of apoptosis proteins in B-cell non-Hodgkin and Hodgkin lymphomas. Cancer. 2006;107:1844–1851. doi: 10.1002/cncr.22219. [DOI] [PubMed] [Google Scholar]
- 28.Rissanen TT, et al. Blood flow remodels growing vasculature during vascular endothelial growth factor gene therapy and determines between capillary arterialization and sprouting angiogenesis. Circulation. 2005;112:3937–3946. doi: 10.1161/CIRCULATIONAHA.105.543124. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda K, Quertermous T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J Biol Chem. 2004;279:55315–55323. doi: 10.1074/jbc.M407776200. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda K, et al. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ Res. 2006;99:424–433. doi: 10.1161/01.RES.0000237662.23539.0b. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda K, et al. Molecular identification and characterization of novel membrane-bound metalloprotease, the soluble secreted form of which hydrolyzes a variety of vasoactive peptides. J Biol Chem. 1999;274:32469–32477. doi: 10.1074/jbc.274.45.32469. [DOI] [PubMed] [Google Scholar]
- 32.Higuchi A, Yamada H, Yamada E, Jo N, Matsumura M. Hypericin inhibits pathological retinal neovascularization in a mouse model of oxygen-induced retinopathy. Mol Vis. 2008;14:249–254. [PMC free article] [PubMed] [Google Scholar]
- 33.Ozaki H, et al. Blockade of vascular endothelial cell growth factor receptor signaling is sufficient to completely prevent retinal neovascularization. Am J Pathol. 2000;156:697–707. doi: 10.1016/S0002-9440(10)64773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo MS, et al. Dramatic inhibition of retinal and choroidal neovascularization by oral administration of a kinase inhibitor. Am J Pathol. 1999;154:1743–1753. doi: 10.1016/S0002-9440(10)65430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.