Embryonic stem (ES) cells and other primitive stem cells of mice have been known for more than 30 years to potently block retrovirus replication1. Infection of ES cells by the murine leukemia viruses (MLVs) results in the normal establishment of integrated proviral DNA, but this DNA is then transcriptionally silenced, preventing further viral spread. The repression is largely mediated by trans-acting factors that recognize a conserved sequence element termed the primer binding site (PBS), an 18-bp sequence complementary to the 3’ end of a cellular tRNA2–6. A specific tRNA is annealed to the PBS sequence of the viral genomic RNA, and is used to prime DNA synthesis7. This same sequence in the context of the integrated proviral DNA is targeted for silencing in ES cells. We recently showed that a large protein complex binding to the PBS in ES cells contains TRIM288,9, a well-characterized transcriptional co-repressor10–12. An important unanswered question is the identity of the factor that directly recognizes integrated retroviral DNAs and recruits TRIM28 to mediate their specific silencing. Here we identify the novel zinc finger protein ZFP809 as the recognition molecule that bridges the integrated proviral DNA and TRIM28. We show that expression of ZFP809 is sufficient to render even differentiated cells highly resistant to MLV infection. We further demonstrate that ZFP809 is able to potently block transcription from DNA constructs of human T-cell lymphotropic virus-1 (HTLV-1), which uses the same primer tRNA. These results identify ZFP809 as a DNA binding factor that specifically recognizes a large subset of mammalian retroviruses and retroelements and targets them for transcriptional silencing. We propose that ZFP809 evolved as a stem cell-specific retroviral restriction factor and therefore constitutes a novel component of the intrinsic immune system of stem cells.
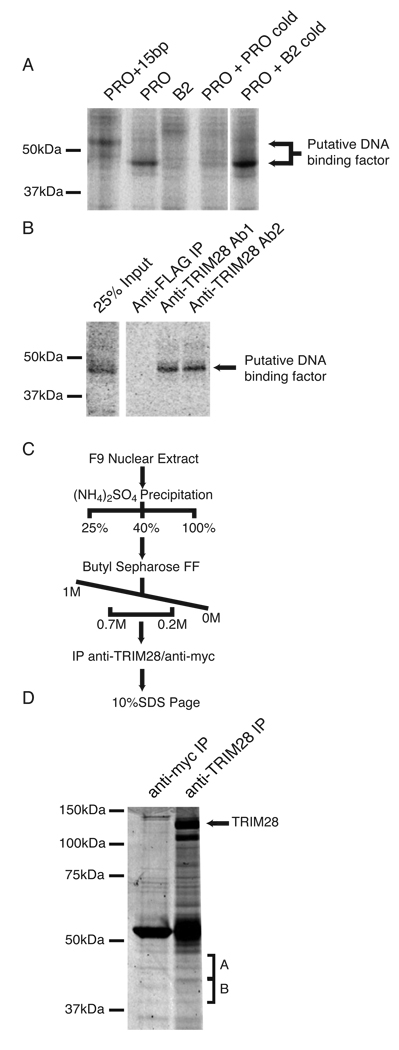
Moloney MLV, a prototypical simple retrovirus, contains a PBS complementary to proline tRNA (PBSPro) and is potently restricted by ES and embryonic carcinoma (EC) cells in a PBS-dependent mechanism. A single bp change known as the B2 mutation (PBSB2) is sufficient to prevent binding of the ES cell silencing factors and so abrogate this restriction2. To identify the DNA-binding component of the complex, we employed a UV-crosslinking approach. We first confirmed that a BrdU-substituted radiolabeled DNA probe corresponding to PBSPro retained the correct specificity for the PBS binding complex (Supplementary Fig. 1). We then incubated this DNA probe with nuclear extracts of the F9 EC cell line, treated the samples with UV light to form covalent protein-DNA crosslinks, and resolved the complexes by SDS-PAGE under denaturing conditions. A crosslinked species migrating at the position of a protein of approximately 45 kDa was observed. This species was not detected when the experiment was performed with a mutant PBSB2 probe, or when a large excess of unlabeled (cold) PBSPro probe was added to the reaction (Fig. 1A). The addition of 15 bp to the PBSPro probe caused the species to migrate at a position corresponding to a mass of ~55 kDa. To confirm that the 45-kDa species was a true component of the PBS binding complex, we showed that this species co-immunoprecipitated with TRIM28, a confirmed subunit of the silencing complex (Fig. 1B). These experiments suggested that a 45-kDa protein was in close contact with DNA and thus was a strong candidate for the sequence-specific recognition factor.
Figure 1. Identification of candidate PBSPro-DNA binding factors.
(A) F9 EC cell nuclear extract prepared and incubated with radiolabeled probes corresponding to PBSPro (PRO), PBSB2 (B2) or PBSPro with a 15 bp extension (PRO+15bp). Crosslinking was performed by exposure to UV light. Competition was achieved through the addition of 100 pmols of unlabeled probe corresponding to either PBSPro or PBSB2. (B) Crosslinked nuclear extracts were subjected to immunoprecipitation with anti-FLAG or two separate anti-TRIM28 antibodies8. (C) Purification scheme used to prepare samples for identification by mass spectrometry. (D) SDS PAGE of samples prepared by purification scheme outlined above, visualized by Coomassie stain. Bands excised for protein identification by mass spectrometry are labeled “A” and “B”.
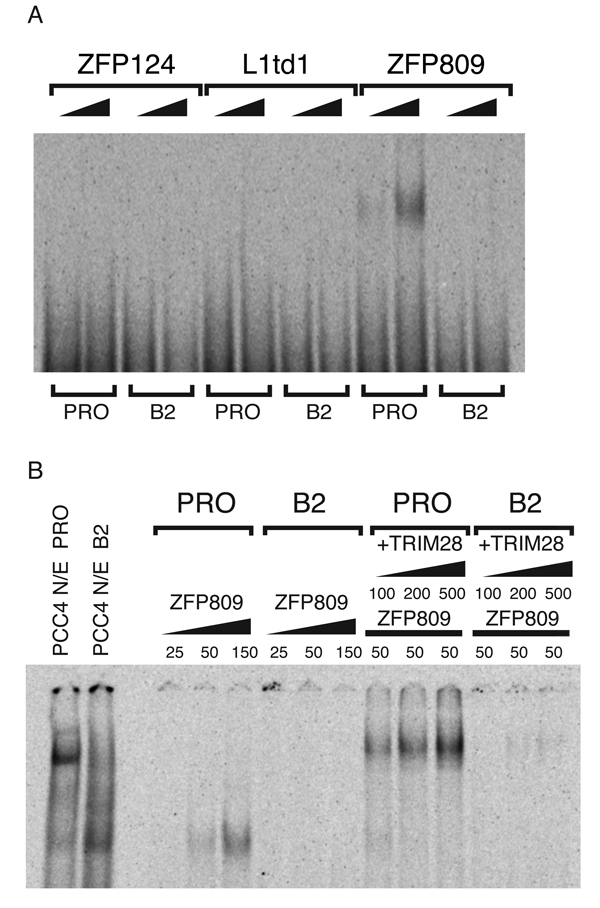
To determine the identity of this protein, we partially purified the PBS silencing complex from F9 EC cell nuclear extracts8 and immunoprecipitated the complex with a TRIM28-specific antibody (Fig. 1C). This immunoprecipitated material was resolved by SDS-PAGE, and bands corresponding t o t h e 4 5-kDa species were excised and characterized by lc-ms-ms mass spectroscopy (Fig. 1D; Supplementary Table 1). Of the proteins identified, the three most likely candidates for DNA-binding factors were L1td1, ZFP809, and ZFP124. L1td1 has sequence similarity to the products of the ORF1 gene of Line-1 retrotransposons, proteins known to bind DNA13. ZFP809 and ZFP124 are both members of the family of KRAB box-containing zinc finger proteins, which both bind DNA 14 and interact with TRIM28 via their KRAB box15,16. To determine which of these candidates has the requisite DNA-binding specificity, we expressed all three in E. coli as recombinant proteins fused to glutathione-S-transferase (GST), and tested lysates in electrophoretic mobility shift assays (EMSAs) with 28-bp probes corresponding to either PBSPro or PBSB2 (Fig. 2A; Supplementary Fig. 2). Of the three candidates, only ZFP809 bound to the WT PBSPro sequence but not to PBSB2, strongly suggesting that it was the DNA recognition component of the PBS silencing complex (Fig. 2A). To determine whether ZFP809 was able to bind simultaneously to PBSPro DNA and to TRIM28, we added increasing amounts of recombinant GST-tagged TRIM28 to EMSA reactions containing ZFP809 and either the PBSPro or PBSB2 probe (Fig. 2B). The addition of TRIM28 dramatically supershifted the ZFP809-PBSPro probe complex, while the PBSB2 probe remained unbound (Fig. 2B). The addition of TRIM28 significantly increased the yield of shifted probe, suggesting that it stabilized the complex or increased the affinity of ZFP809 for the PBSPro probe. Further, the PBS binding complex that formed in the presence of both recombinant TRIM28 and ZFP809 proteins exhibited virtually identical migration to the PBS complex formed in nuclear extracts from EC cells, suggesting that these two components alone are sufficient to reconstitute the minimal PBS silencing complex (Fig. 2B). Furthermore the DNA binding specificity of this recombinant reconstituted complex was found to be virtually indistinguishable from the native complex (Supplementary Fig. 3).
Figure 2. Reconstitution of the PBSPro binding complex with recombinant proteins.
(A) 50 ng or 250 ng of recombinant GST-tagged ZFP124, L1td1 and ZFP809 added to EMSA reactions with radiolabeled labeled probes corresponding to PBSPro (PRO) or PBSB2 (B2). (B) EMSAs performed as above, with PCC4 EC nuclear extract or 25, 50, or 100 ng of GST-ZFP809 protein and 100 ng, 200 ng or 500 ng of GST-TRIM28 of protein as indicated.
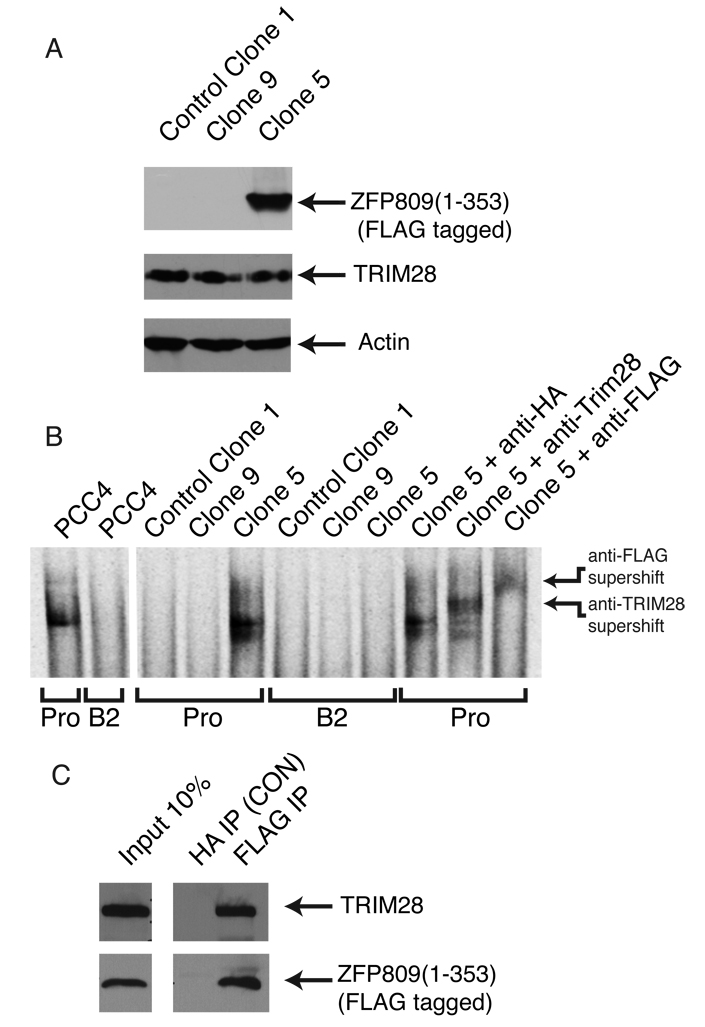
These experiments strongly suggested that ZFP809 was the DNA recognition component of the PBS silencing complex, and because TRIM28 is ubiquitous and present at high concentrations, suggested the possibility that ZFP809 was also the critical limiting component in ES cells and thus sufficient to induce virus resistance. We therefore asked whether expression of this protein in a permissive cell line could render it resistant to MLV or other viruses utilizing a PBS specific for proline tRNA. Initial attempts to generate 293A cell lines stably expressing full-length ZFP809 were unsuccessful and most often yielded clones with 3’ deletions, suggesting that the C-terminal 50 residues are toxic in non-stem cells. Subsequent experiments were performed with constructs expressing the ZFP809 protein truncated at amino acid 353 (ZFP809(1-353)), which showed minimal toxicity and was expected to retain its silencing functionality as it included both DNA-binding and TRIM28-binding domains (Supplementary Fig. 4). 293A cells stably expressing ZFP809(1-353) with an N-terminal FLAG tag were generated and individual clones were screened for expression of ZFP809(1-353) (Supplementary Fig. 5B). Clones expressing the empty vector were constructed in parallel. Three representative cloned lines were selected for further study: Clone 5, which expressed representative levels of ZFP809(1-353); Clone 9, which had been transformed with the ZFP809(1-353) DNA but failed to express ZFP809; and control Clone 1, which contained the empty vector (Fig. 3A and Supplementary Fig 5B). The cell lines were all confirmed to retain normal TRIM28 expression (Fig. 3A). Nuclear extracts were prepared and tested for the presence of the silencing complex by EMSAs. The line expressing ZFP809(1-353) exhibited a strong PBSPro binding activity (Fig. 3B). This reconstituted PBS silencing complex showed the same specificity as the native EC cell complex and did not bind to the PBSB2 probe (Fig. 3B). The shifted complex produced by expression of ZFP809(1-353) in 293A cells was shown to contain both endogenous TRIM28 and exogenous FLAG-tagged ZFP809(1-353), as judged by supershifts with anti-TRIM28 and anti-FLAG antibodies. The interaction between ZFP809 and TRIM28 in vivo was confirmed by co-immunoprecipitation in Clone 5 cells (Fig 3C)
Figure 3. Stable expression of ZFP809 in 293A cells leads to reconstitution of the PBS silencing complex in vivo.
(A) Nuclear extracts from clonal cell lines control Clone 1 (empty vector control), Clone 5 (cell line stably expressing FLAG-tagged ZFP809(1-353) or Clone 9 (cell line generated with same construct as Clone 5 but which does not express ZFP809) probed by western blot with antisera for FLAG (upper panel), TRIM28 (middle panel), or beta-actin (lower panel). (B) Nuclear extracts prepared either from PCC4 EC cells or cell lines control Clone 1, Clone 9, or Clone 5 used in EMSA reactions with radiolabeled probes corresponding to PBSPro (PRO) or PBSB2 (B2). In final three lanes the PBS silencing complex is supershifted by the addition of 2 µg of either anti-TRIM288, anti-FLAG, or a control anti-HA antibody. (C) ZFP809(1-353) and TRIM28 co-immunoprecipitate in DNase treated Clone 5 whole cell extracts in the presence of ethidium bromide. Control immunoprecipitations were performed with of HA antisera. Immunoprecipitated ZFP809(1-353) and Co-immunoprecipitated TRIM28 was detected by western blot.
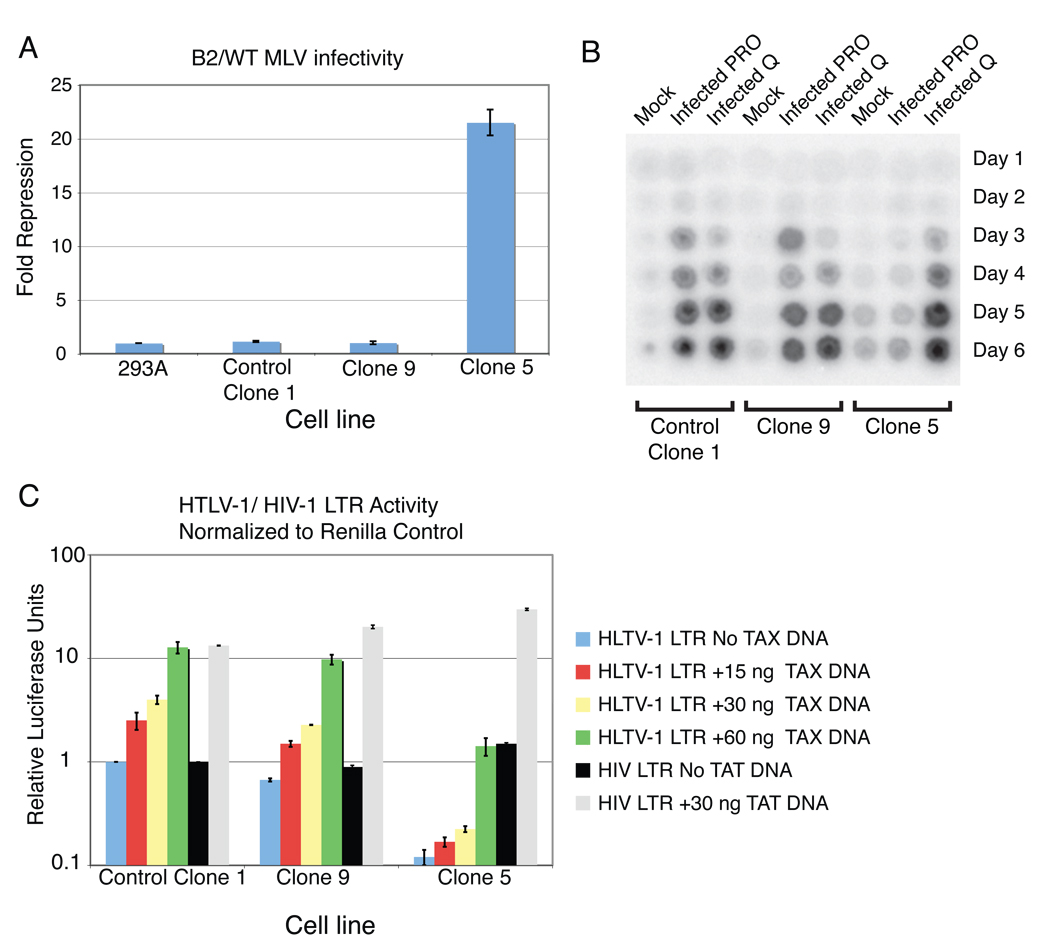
To test whether reconstitution of the minimal PBS silencing complex was sufficient to induce silencing of MLV in a PBS-dependent manner, we used two MLV reporter constructs identical except for a single base pair change in the PBS sequence. MLV particles pseudotyped by the VSV G protein, transducing the puromycin resistance gene, and utilizing either PBSPro or PBSB2, were generated. The infectivity of these two virus preparations was determined by colony formation assays after infection of the Clone 5, and the controls Clone 1, Clone 9, and the parental 293A cell lines. The ratio of infectivity of the PBSB2 MLV over that of the PBSPro MLV in a given cell line is a measure of the level of PBS-mediated silencing, after normalizing the ratio for the parental cell line to 1. The control Clone 1 and Clone 9 cell lines had a ratio close to 1, indicating that they exhibited no PBS-mediated restriction. In contrast, Clone 5 demonstrated potent PBS-mediated silencing, manifesting as a ratio of ~23 fold (Fig. 4A). Analysis of other clones expressing ZFP809(1-353) showed that all exhibited potent PBS-mediated silencing (Supplementary Fig 5A). Thus, expression of ZFP809(1-353) was sufficient to render a differentiated cell resistant to transduction by PBSPro-utilizing retroviral vectors. To investigate whether ZFP809 silences retroviral expression by binding directly to the integrated provirus, leading to the recruitment of TRIM28, we performed chromatin immunoprecipitations on lysates of clone 5 cells infected either with a restricted (PBSPro) or unrestricted variant PBS corresponding to a glutamine tRNA (PBSQ) MLV (Supplementary Fig 6). We found that both ZFP809 and TRIM28 are significantly enriched at PBSPro proviral sites (Supplementary Fig 6).
Figure 4. Expression of ZFP809 in a differentiated cell line causes a potent block to the replication of PBSPro utilizing retroviruses.
(A) 293A, control Clone 1, Clone 9 and Clone 5 cell lines were infected with either PBSPro or PBSB2 VSV-G pseudotyped MLV virions expressing the puromycin resistance gene. Infection efficiency in each cell line was monitored by colony count after of puromycin selection. Graph shows ratio of B2/PRO infection efficiency in each cell line, normalized with 293A =1. Error bars show +/− standard error, with n=3. (B) Control Clone 1, Clone 9 and Clone 5 cell lines were infected at a low multiplicity with replication-competent amphotropic MLV utilizing either a PBSPro (PRO) or PBSQ (Q). Viral spread in these cell lines was monitored by analysis or reverse transcriptase activity22 in the culture media every day for 6 days. (C) HTLV-1 LTR or HIV-1 LTR firefly luciferase reporter activity monitored in cell lines control Clone 1, Clone 5, and Clone 9 which were simultaneously transfected with increasing amounts of a Tax expressing vector (pTAX) or TAT expressing vector (pTAT-HA) as shown. Firefly luciferase values normalized to renilla luciferase values from a simultaneously transfected renilla control vector. Error bars show +/− standard error, with n=3.
To assess whether ZFP809 could also reduce virus replication after authentic retrovirus infection we generated amphotropic MLV virus constructs containing either the restricted PBSPro or unrestricted PBSQ. These viruses were used separately to infect the Clone 5 and control Clone 1 and Clone 9 cell lines at a low multiplicity of infection, and productive spread of the virus was monitored by measurement of reverse transcriptase activity in the culture media. The amphotropic virus using the wild-type PBSPro could replicate in the control Clone 1 and Clone 9 cell lines, but was completely blocked in the Clone 5 cell line (Fig. 4B). The amphotropic MLV using PBSQ was able to spread in all the cell lines.
To investigate whether ZFP809 expression is required for PBS mediated restriction in embryonic cells, ZFP809 expression was attenuated in F9 EC cells by RNAi. Substantial knockdown of ZFP809 correlated with a complete relief of PBS mediated restriction (Pools 8,9 and 12) (Supplementary Fig 7). mRNA analysis also showed lower expression of ZFP809 in non-restrictive NIH3T3 cells when compared to restrictive ES or F9 cells, consistent with the hypothesis that that there is a threshold level of ZFP809 required for PBS mediated restriction (Supplementary Fig 7).
Having shown that MLV is potently restricted by ZFP809, we wished to determine whether the human pathogen HTLV-1, which also utilizes a PBSPro, would also be restricted. A complication to this question is that unlike MLV, HTLV-1 expresses the accessory protein Tax, which recruits co-activators to the LTR and stimulates transcription17,18. It is therefore conceivable that Tax-mediated stimulation of transcription from the LTR might overcome the block induced by ZFP809. To score for transcriptional silencing, we utilized an HTLV-1 LTR firefly luciferase reporter construct co-transfected with increasing amounts of a Tax expressing plasmid, and with a TK-renilla luciferase control plasmid (for normalization) into Clone1, Clone 9 and Clone 5 cell lines (Fig. 4C). The HIV-1 LTR (which does not contain a PBSPro) and the coexpression of the Tat transactivator protein were also tested, as a negative control (Fig. 4C). In the case of basal transcription in the absence of Tax, the HTLV-1 LTR reporter construct was silenced 8 fold in Clone 5 when compared to control Clone 1, and with increasing amounts of Tax this fold silencing was increased, with Clone 5 having on average a 14-fold lower LTR activity when compared with control Clone 1 (Fig. 4C). These results demonstrate that the HTLV-1 LTR is restricted, and that Tax expression does not abrogate ZFP809-mediated restriction. In order to demonstrate that ZFP809 mediated restriction requires TRIM28, its levels were stabily reduced in Clone 5 by RNAi (Supplementary Fig 8). TRIM28 knockdown resulted in a decrease in PBS binding activity in Clone 5 nuclear extracts and a reduction in HTLV-1 LTR restriction in these cells (Supplementary Fig 8). We note that TRIM28 knockdown in Clone 5 cells also led to a decrease in ZFP809 levels suggesting that ZFP809 protein stability is reliant on formation of the TRIM28-ZFP809 complex (Supplementary Fig 8).
These findings indicate that ZFP809 is the critical DNA-binding factor in ES cells that recognizes integrated PBSPro-utilizing retroviral DNAs and silences them through the recruitment of TRIM28. We have shown that viruses utilizing the distinctive Lys1,2 tRNA are also silenced in stem cells in a TRIM28-dependent manner19, and thus that there must exist a factor recognizing this distinctive PBS. We speculate that ZFP809 represents the first in a class of PBS-specific retroviral restriction factors that all act through the recruitment of TRIM28 to different PBS elements to silence integrated retroviruses and thereby act as components of a stem cell-specific intrinsic immunity.
Methods Summary
Electrophoretic mobility shift assay (EMSA)
Double-stranded DNA probes were 5’ end-labeled using [γ-33P] ATP. Binding reactions were performed using Modified Thornell binding buffer8 (25 mM HEPES [pH 7.9], 1 mM EDTA, 10% [v/v] glycerol, 5 mM DTT, 25 ng poly(dI-dC) per µl, 5 mM NaCl, 5 mM KCl, 3 mM MgCl2, and 0.1 mM ZnCl2). Probes (50,000 CPM) were incubated with nuclear extract for 25 min at 30°C in a total volume of 20 µl. For supershifts and cold DNA competitions, antibody/cold DNA was added at same time as probe. Binding reactions were analyzed by electrophoresis on 4% native polyacrylamide gels8.
UV crosslinking
Proteins in F9 nuclear extracts were resuspended in Modified Thornell binding buffer8 and 150000cpm 33P labeled probe added to each 50 µl reaction. Samples were then exposed to 302 nm UV light for 45 minutes using a transilluminator (VWR). Competitions were performed with the addition of 100 pmols of unlabeled probe per reaction.
Co-Immunoprecipitations
107 Clone 5 cells were lysed in 750 µl IPH buffer21 (150 mM NaCl, 50 mM Tris-HCl, pH 8, 0.5% Nonidet P-40, 5 mM MgCl2 1 mM CaCl2 0.5 mM EDTA, + protease inhibitors [Complete, Roche]). The lysates were then treated with 100 U DNase (Roche) for 24 h at 4°C. Ethidium bromide was subsequently added to lysates (50 µg/ml) and co-immunoprecipitations were performed using 5 µg of antibody and a mixture of 20 µl protein A and 20 µl protein G Dynabeads (Invitrogen).
Retroviral transduction assays
Virus stocks were harvested as previously described8. Retroviral preparations were then serially diluted (for titer determination), and added to F9, NIH3T3 or 293A cells (seeded at [F9] 3.5 × 103 and [NIH3T3/293A] 2 × 103 cells per cm2 the day prior to transduction) in the presence of 8 µg of Polybrene/ml. G418 or puromycin containing selective media was added 24–48 h post infection and colonies were counted 14 days after selection.
LTR reporter assays
293A cells were seeded 7000 cells per well in a 96 well plate and were transfected using Fugene 6 (Roche) with 20 ng HTLV-1 LTR or HIV-1 LTR firefly luciferase reporter plasmid, 5ng of a TK promoter renilla luciferase control plasmid (phRL-tk, Promega), and a Tax or TAT expressing vectors. Luciferase activity was assayed 48 hours post infection using dual-luciferase reporter assay (Promega).
Supplementary Material
Acknowledgements
This work was supported by PHS grant R37 CA 30488 from the National Cancer Institute. We thank David Derse, Fadila Bouamr and Eric Barklis for their generosity with reagents. We are grateful to Martha de los Santos, Helen Nickerson and Mary Ann Gawinowicz (Protein Core Facility Columbia University) for technical assistance. We would also like to thank Drs Adusumilli, Banes, Bilsky Yamada and co-workers, without whose help this paper would never have been written. DW is an Associate, and SPG is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Teich NM, Weiss RA, Martin GR, Lowy DR. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12(4):973. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 2.Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47(3):391. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 3.Feuer G, Taketo M, Hanecak RC, Fan H. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J Virol. 1989;63(5):2317. doi: 10.1128/jvi.63.5.2317-2324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11(3):1214. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi M, et al. Stem cell factor binding to retrovirus primer binding site silencers. J Virol. 1995;69(2):1142. doi: 10.1128/jvi.69.2.1142-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf D, Goff SP. Host Restriction Factors Blocking Retroviral Replication. Annu Rev Genet. 2008;42:143. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harada F, Peters GG, Dahlberg JE. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem. 1979;254(21):10979. [PubMed] [Google Scholar]
- 8.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131(1):46. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Wolf D, Cammas F, Losson R, Goff SP. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol. 2008;82(9):4675. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayyanathan K, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17(15):1855. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Geen H, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3(6):e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuruma R, et al. Physical and functional interactions between STAT3 and KAP1. Oncogene. 2008;27(21):3054. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 13.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A. 1997;94(19):10155. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271(5252):1081. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 15.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4(10):231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman JR, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 17.Felber BK, et al. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science. 1985;229(4714):675. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 18.Bex F, Gaynor RB. Regulation of gene expression by HTLV-I Tax protein. Methods. 1998;16(1):83. doi: 10.1006/meth.1998.0646. [DOI] [PubMed] [Google Scholar]
- 19.Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci U S A. 2008;105(34):12521. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuks F, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278(6):4035. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 21.Wolf D, et al. Acetylation of beta-catenin by CREB-binding protein (CBP) J Biol Chem. 2002;277(28):25562. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- 22.Telesnitsky A, Blain S, Goff SP. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.